- Home
- News & Updates
- Accelerating Research of SARS-CoV-2 Host Response
-
News
- 07/08/2020
Accelerating Research of SARS-CoV-2 Host Response
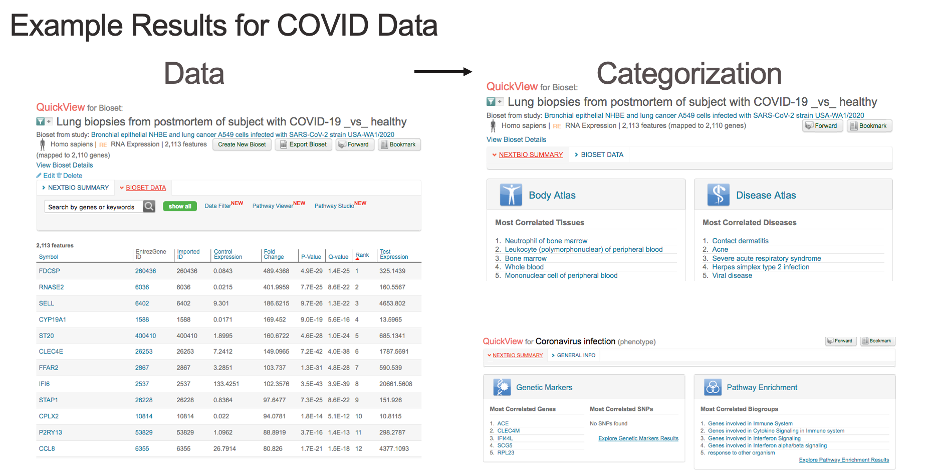
As scientists seek to address the COVID-19 pandemic, the study of the human host response to SARS-CoV-2 plays a critical role. It is vital to understand why some individuals become severely ill, what biomarkers may warn of this outcome, what pathways play important roles, and what drugs might represent possible leads for treatment.
Earlier this morning, we released a new collaborative environment to the COVID-19 research community to enable and accelerate SARS-CoV-2 host response research leveraging Illumina’s BaseSpace Correlation Engine. Available to the COVID-19 community free-of-charge for 6 months, the new Correlation Engine environment provides a dynamic ecosystem for researchers to put their results into context, accelerating discoveries and enabling hypothesis validation around host responses, biomarker identification and drug discovery.
The environment contains the full suite of Correlation Engine tools, combined with several pre-processed data sets based on coronavirus animal model systems, as well as recently released public SARS-CoV-2 data. Additional scientific data will be provided into the resource as it becomes available. The environment will enable interested researchers to discover important pathways, biomarkers, and potential drug candidate leads, to help address the COVID-19 crisis.
The resource will be made available free-of-charge for 6 months to SARS-CoV-2 researchers at https://covid-19.ussc.informatics.illumina.com/c/; they are encouraged to apply for access by contacting Illumina directly via the Illumina COVID response website, or via their Illumina rep. Although sharing data is not a requirement, researchers are encouraged to submit datasets for addition to the resource, or highlight public datasets that they believe should be added to advance the cause of understanding SARS-CoV-2 host response. The platform supports results generated from Illumina’s sequencing and array platforms, as well as complimentary data from other experimental methodologies. A variety of datasets can be imported into the BSCE system for correlation analysis, including gene expression data, methylation, GWAS, protein interaction, and more.
To see examples of how BSCE may be leveraged to better understand SARS-CoV-2 host response, interested researchers can review a series of videos created by BSCE users Mike Edwards, Ph.D., Enrique Torchia, Ph.D., and collaborators at: https://www.youtube.com/playlist?list=PLCQx4mwID7jtl-nDsH-aKktNp_zNV7zle Data developed in these videos has been made available in the resource.
This new resource for researching SARS-CoV-2 host response complements capabilities for laboratory viral sequencing workflows, and viral genomics, and viral surveillance, and testing recently announced by Illumina.
The SARS-CoV-2 collaborative BSCE environment is for research use only.
For research use only. QB #1033