- Home
- News & Updates
- BaseSpace™ Sequence Hub Run Planner now includes 10x Genomics libraries.
-
BaseSpace™ Sequence Hub
-
Product updates
-
News
- 02/19/2024
BaseSpace™ Sequence Hub Run Planner now includes 10x Genomics libraries.
The Run Planning tool can be used to set up a sequencing run in BaseSpace Sequence Hub (BSSH). We’re excited to announce that sequencing runs with 10x Genomics libraries can now be directly set up on the Run Planner.
What is the run planner tool?
Once logged into BaseSpace Sequence Hub (BSSH), the Run Planner tool guides you through the steps needed for preparing your sequencing run, such as specifying sample and index information, read length and analysis options. After saving, the planned run can directly be accessed and selected on your instrument. If your instrument is not connected to the cloud, you can use Run Planning to generate a sample sheet v2 file to configure a local sequencing run.
Planning a run in BaseSpace Sequence Hub is available for the following sequencing systems:
- NextSeqTM 1000 and NextSeq 2000 Sequencing Systems
- NovaSeqTM X Series Sequencing Systems
- NovaSeq 6000 Sequencing System
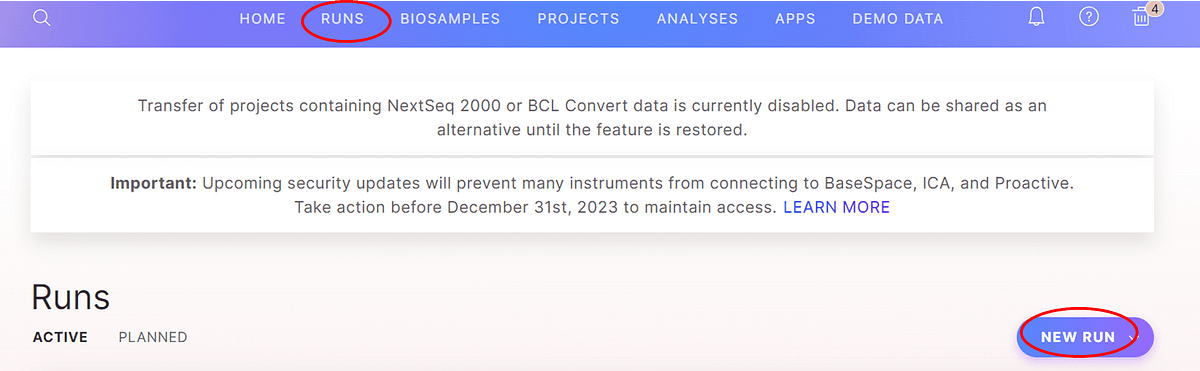
Figure 1: Access the run planner by selecting “runs” and then “new run” on the BSSH page. Then select “run planning” tool.
Select the 10x Genomics library from the drop down menu
Illumina and 10x Genomics team have collaborated to include 10x Genomics single cell and spatial libraries on the Run Planner tool. All on-market 10x Chromium and Visium products are currently available in the Illumina's BSSH Run Planner. Please refer to the following Direct Demultiplexing pages on the 10x Genomics Support site, linked below:
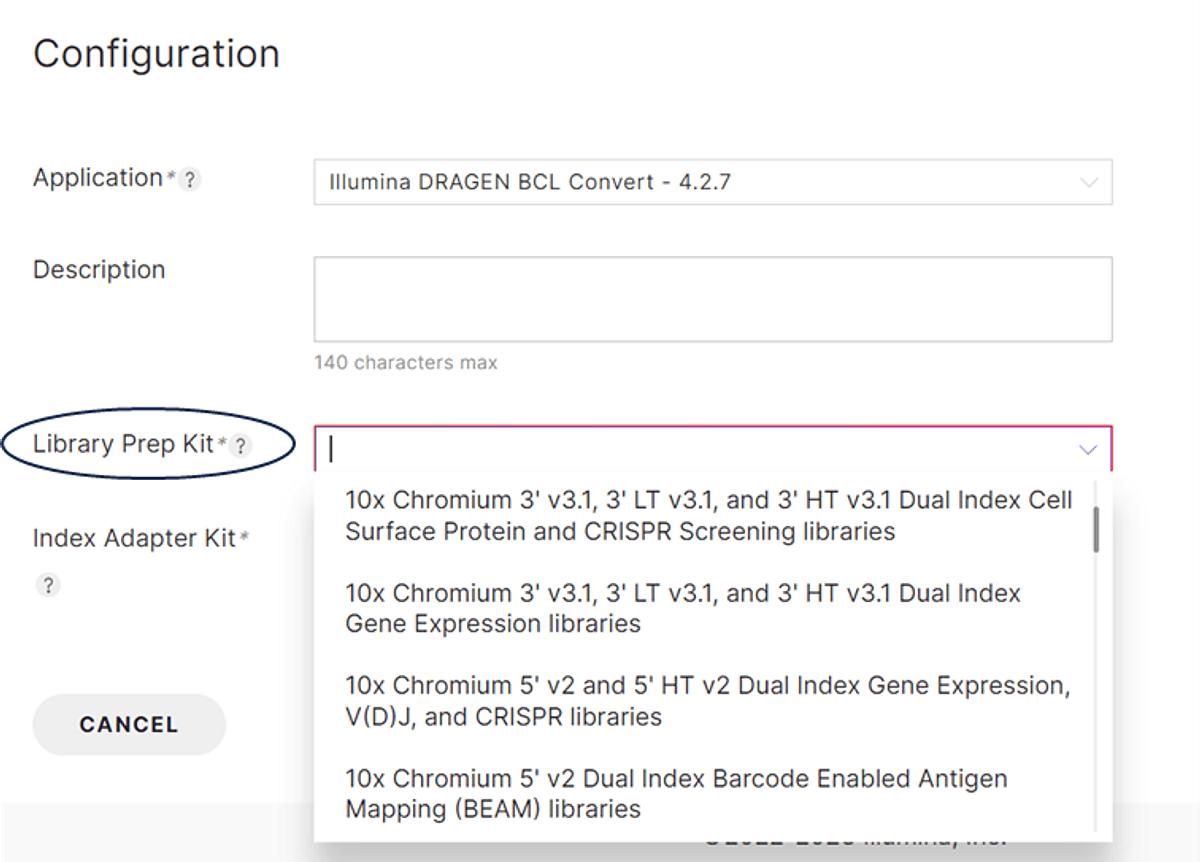
Figure 2: Selection of the 10x Genomics library prep kit on the Run Planner tool. Use the drop down menu to select the library prep kit of choice.
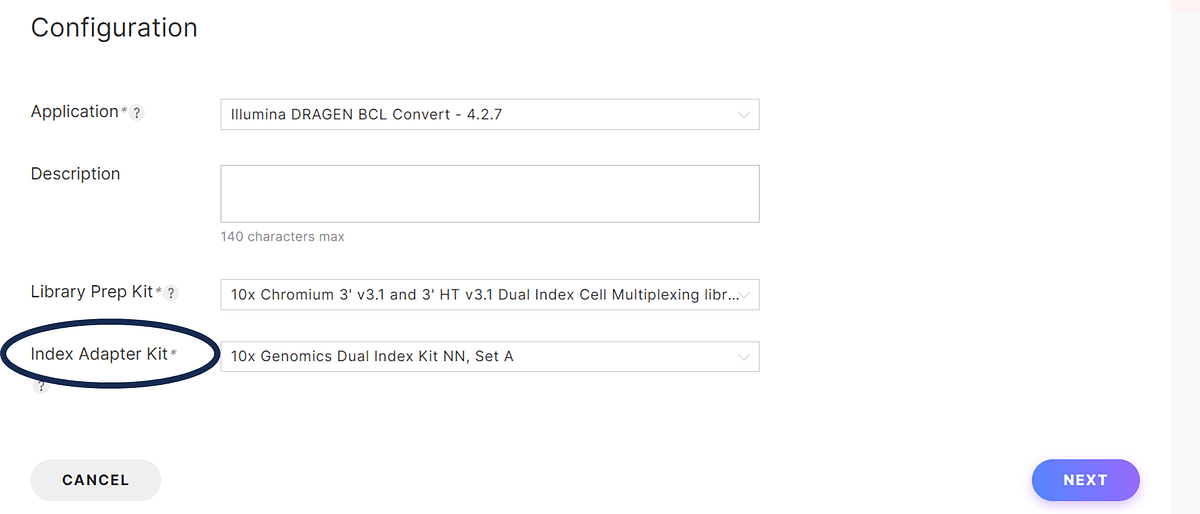
Figure 3: Selection of the 10x Genomics index kit on the Run Planner tool. Use the drop down menu to select the index kit of choice.
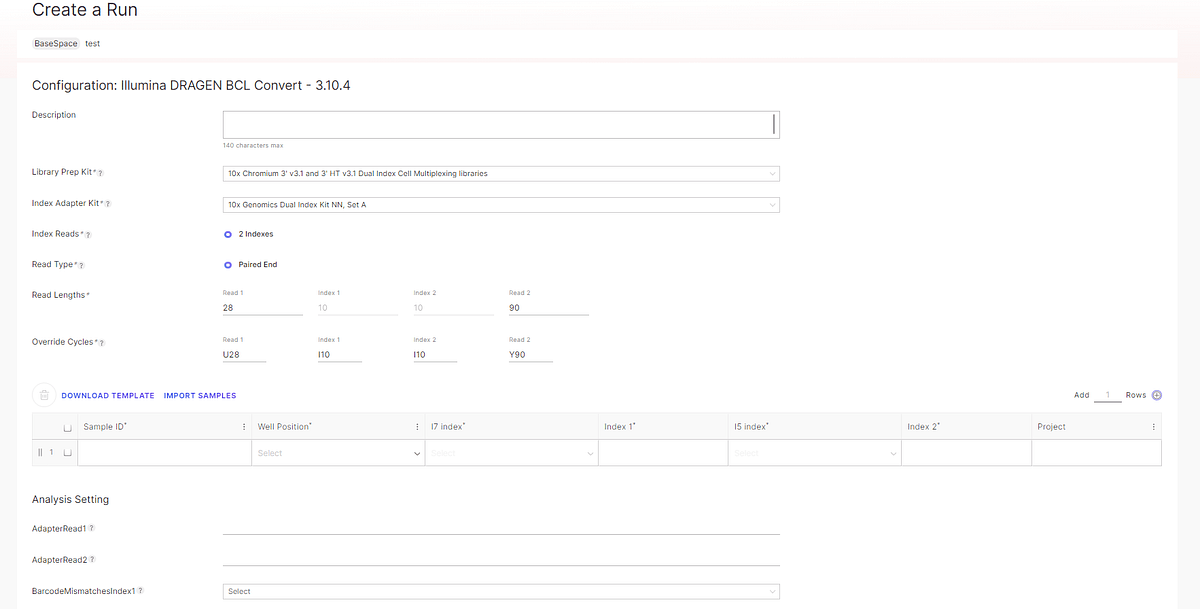
Figure 4: run creation. Selecting the 10x Genomics library prep kit and index will autopopulate read length and index scheme. The 'Library Prep Kit' and 'Index Adapter Kit' are automatically populated by your selections on the previous page. The Read Lengths and Override Cycles are also automatically populated. You do not need to change these – but it is possible to change the Read Lengths if necessary.
Please make sure to leave the AdapterRead1 and AdapterRead2 entries blank. If adapter trimming is done during demultiplexing, there is a risk of losing barcode and UMI information in downstream analysis. We generally do not recommend any adapter trimming or preprocessing FASTQ reads for any 10x product.
You can also leave the BarcodeMismatchesIndex1 and BarcodeMismatchesIndex2 blank. This will allow the default setting of 1 mismatch.
Make sure 'gzip' is selected for FASTQ Compression Format. Then click Next to go to the next page. Next step is to save the run. Once the run has been saved, it is possible to export the sample sheet as a csv. file.
Support
For further questions regarding the Run planner, please contact Illumina at techsupport@illumina.com
For any questions regarding the libraries, please contact 10x Genomics at support@10xgenomics.com
References
https://support-docs.illumina.com/IN/CloudRunSetup/Content/IN/NovaSeqX/PlanCloudHybridRun.htm
https://help.basespace.illumina.com/sequence/plan-runs
https://help.basespace.illumina.com/sequence/plan-runs/multi-analysis-planned-run
https://help.basespace.illumina.com/sequence/plan-runs/set-up-planned-run
For Research Use Only. Not for use in diagnostic procedures. M-GL-02657