- Home
- News & Updates
- BioLegend BEN-seq libraries sequencing runs now available on Illumina® BaseSpace™ Sequence Hub
-
BaseSpace™ Sequence Hub
-
News
- 07/21/2022
BioLegend BEN-seq libraries sequencing runs now available on Illumina® BaseSpace™ Sequence Hub
Today, we’re sharing the sixth in a series of posts that shed more light on new run data publicly available on BaseSpace™ Sequence Hub (BSSH).
Recent developments in multiomics sequencing technologies have combined the single-cell analysis of RNA and protein in a technique called cellular indexing of transcriptomes and epitopes by sequencing (CITE-Seq). This method uses antibodies conjugated to oligonucleotides, available as TotalSeq antibodies from BioLegend. These oligo-conjugated antibodies have since been incorporated into a bulk RNA-Seq workflow for simultaneously obtaining RNA and protein data in the absence of a single-cell instrument using Illumina next-generation sequencing (NGS), described as bulk epitope and nucleic acid sequencing (BEN-Seq), rendering a complete proteogenomic profile of the sample.
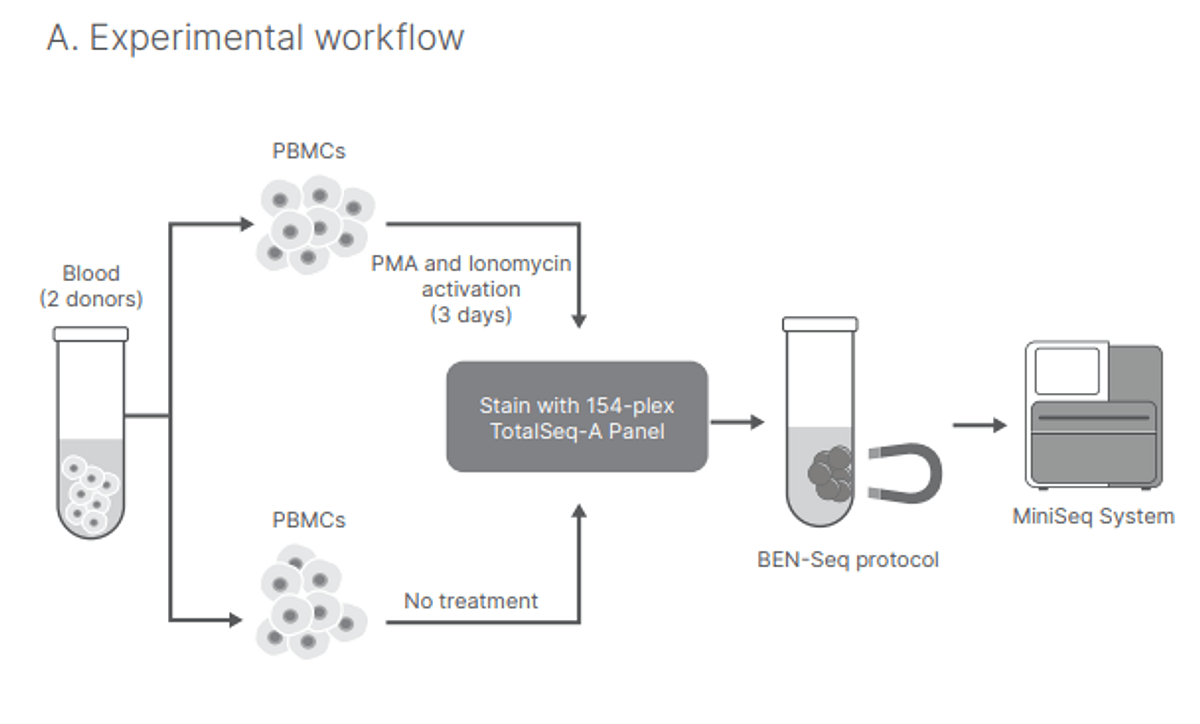
Figure 1 – Experimental workflow
Human PBMCs were isolated from healthy donors. To incorporate protein analysis into bulk RNA-Seq, PBMCs for BEN-Seq were stained with 154 TotalSeq-A antibodies comprising the TotalSeq-A Human Universal Cocktail (https://www.biolegend.com/en-us/products/totalseq-a-human-universal-cocktail-v1-0-20321), and using the BioLegend BEN-Seq protocol. RNA and antibody-derived tag (ADT) fractions of PBMCs were isolated and separated using streptavidin-conjugated magnetic beads loaded with complementary oligonucleotides following the BioLegend BEN-Seq protocol. The remaining RNA fraction was processed using the Illumina TruSeq™ Stranded mRNA Library Prep Kit with Illumina TruSeq RNA UD Indexes.

Table 1: Input requirements for bulk-cell proteogenomics samples and recommended number of sequencing reads
Bulk-ADT samples were sequenced at BioLegend on Illumina MiniSeq™ high output flowcell with a 30 + 8 + 30 run (https://support.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/biolegend-ben-seq-app-note-m-gl-00024/biolegend-ben-seq-app-note-m-gl-00024.pdf) and analyzed with a command-line script using Kallisto and DESeq2.
Run 1: https://basespace.illumina.com/s/cTXOGRbgdY6q
Run 2: https://basespace.illumina.com/s/MLknbkxSqI9E
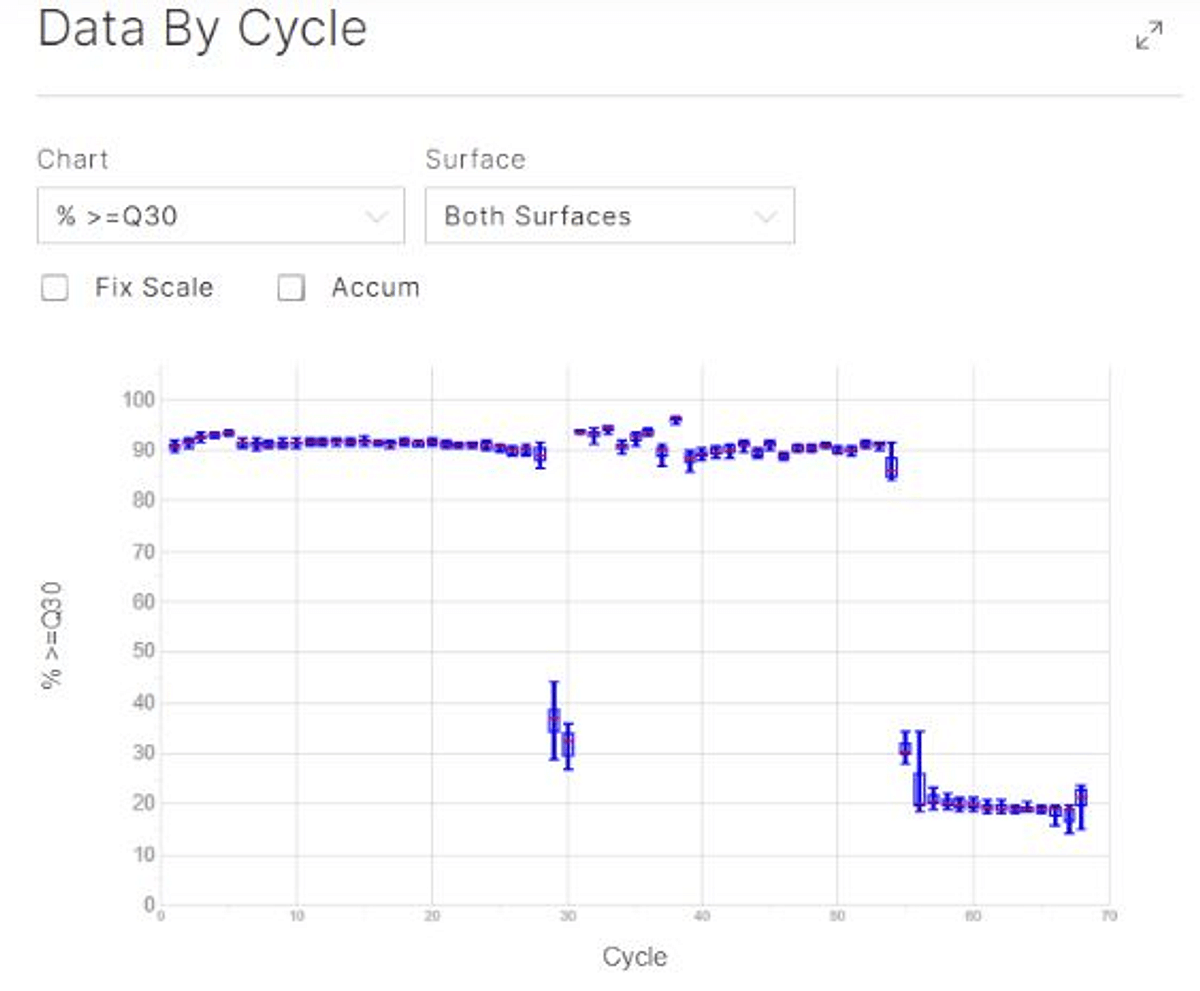
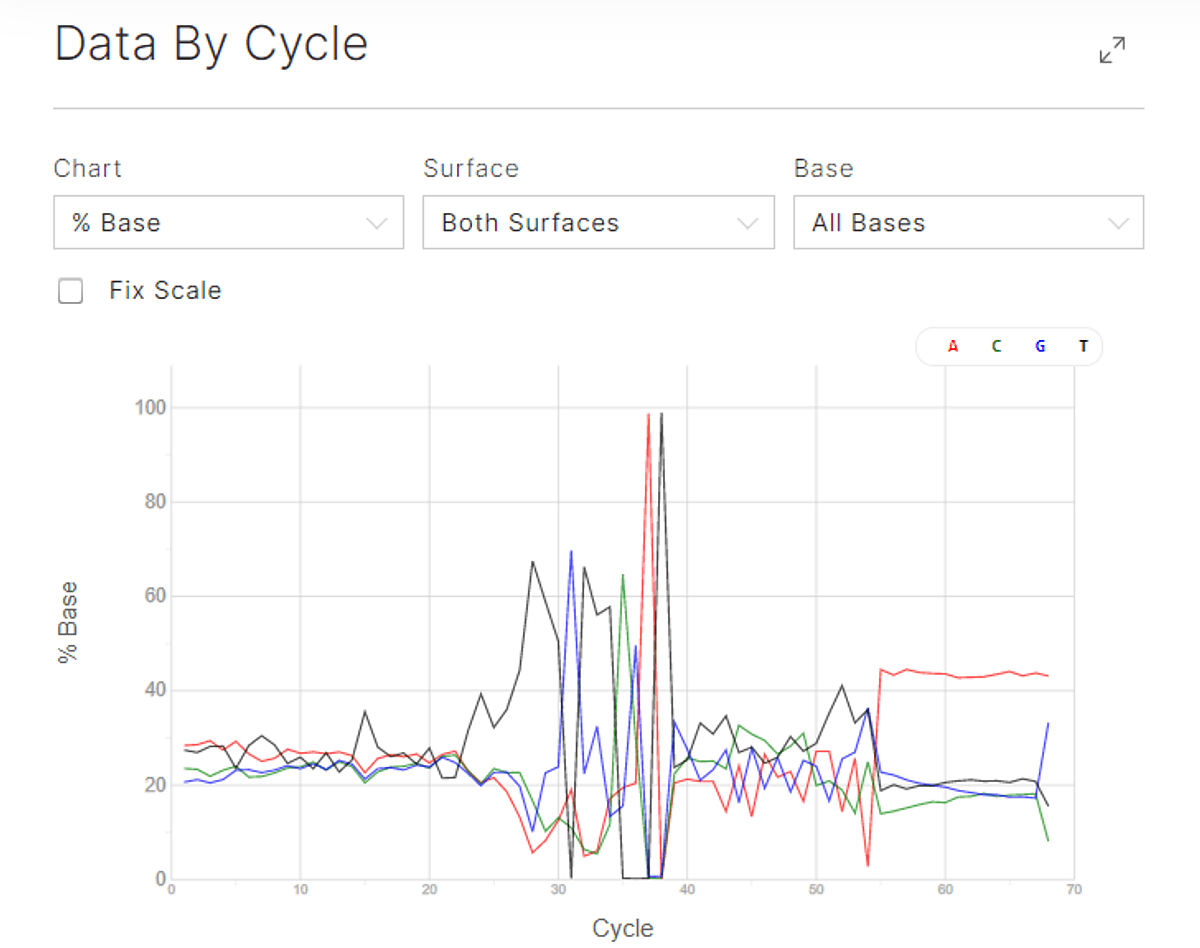
Figure 3: The BEN-seq libraries have very short inserts, especially in the reverse reads, resulting in an A-overcall and expected Q30 drop after 55 cycles of sequencing.
No PhiX was spiked in in these runs, but it is possible to spike in 1% PhiX as a quality control of the sequencing runs with BioLegend BEN-seq libraries.
Additional resources:
- https://www.biolegend.com/en-us/totalseq?gclid=Cj0KCQjwnNyUBhCZARIsAI9AYlEQLRfJqipvEJw41tStTZNt1ra9HOra4AOzU3AZCX9gjl1owrlqHnUaArnSEALw_wcB
- https://www.biolegend.com/en-us/protocols/bulk-epitope-and-nucleic-acid-sequencing-ben-seq-protocol
- https://support.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/biolegend-ben-seq-app-note-m-gl-00024/biolegend-ben-seq-app-note-m-gl-00024.pdf
- https://www.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/biolegend-totalseq-dilution-application-note-m-gl-00475/biolegend-totalseq-dilution-application-note-m-gl-00475.pdf
Support
Contact BioLegend for any questions regarding the antibodies, the library prep assay and the downstream analysis. Contact Illumina Tech support for sequencing related questions.
Products mentioned here are for research use only and are not for diagnostic use.
Special thanks to our colleagues at BioLegend for generating and sequencing BEN-seq libraries on their MiniSeq instrument.
For Research Use Only. Not for use in diagnostic procedures.