- Home
- News & Updates
- BioLegend Cell Hashing/TotalSeq libraries sequencing runs now available on Illumina® BaseSpace™ Sequence Hub
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 04/27/2023
BioLegend Cell Hashing/TotalSeq libraries sequencing runs now available on Illumina® BaseSpace™ Sequence Hub
Today, we are sharing more data in a series of posts that shed more light on new run publicly available on BaseSpace™ Sequence Hub (BSSH).
CITE-seq, or Cellular Indexing of Transcriptomes and Epitopes by Sequencing is a single-cell analysis technique in which cells are stained with antibodies conjugated to short DNA oligos. These oligos contain a sequence barcode, also known as an antibody derived tag (ADT), which is used to identify targeted cellular surface epitopes. These oligo-conjugated antibodies are available as TotalSeq antibodies from BioLegend. Protein and RNA expression can be characterized in single cells with the use of TotalSeq antibodies in conjunction with the scRNA-seq workflow. CITE-Seq can also be paired with cell hashing to allow sample multiplexing during sequencing. Please note that CITE-seq based cell hashing/ multiplexing is different that the multiplexing of individual sequencing libraries that Illumina enables using indexing of the sequenced reads. See our CITE-seq webpage for more details: https://www.illumina.com/techniques/sequencing/rna-sequencing/cite-seq.html
Cell hashing is a technique used to stain cells for multiplexing single-cell partitioned samples during sequencing. Each cell hashing reagent contains a mixture of two distinct monoclonal antibodies targeting distinct ubiquitous cellular surface epitopes. Each monoclonal antibody is conjugated with a short DNA oligo containing the same sequence barcode, also known as hashtag oligo (HTO). HTOs are used to correlate tagged cells with their sample of origin, and they can be mixed together after staining. Cell hashing is a CITE-seq application and therefore is compatible with a standard CITE-seq experiment, as well as with and other single-cell analysis techniques. Cell Hashing is a method that enables sample multiplexing and super-loading on single cell RNA-sequencing platforms.
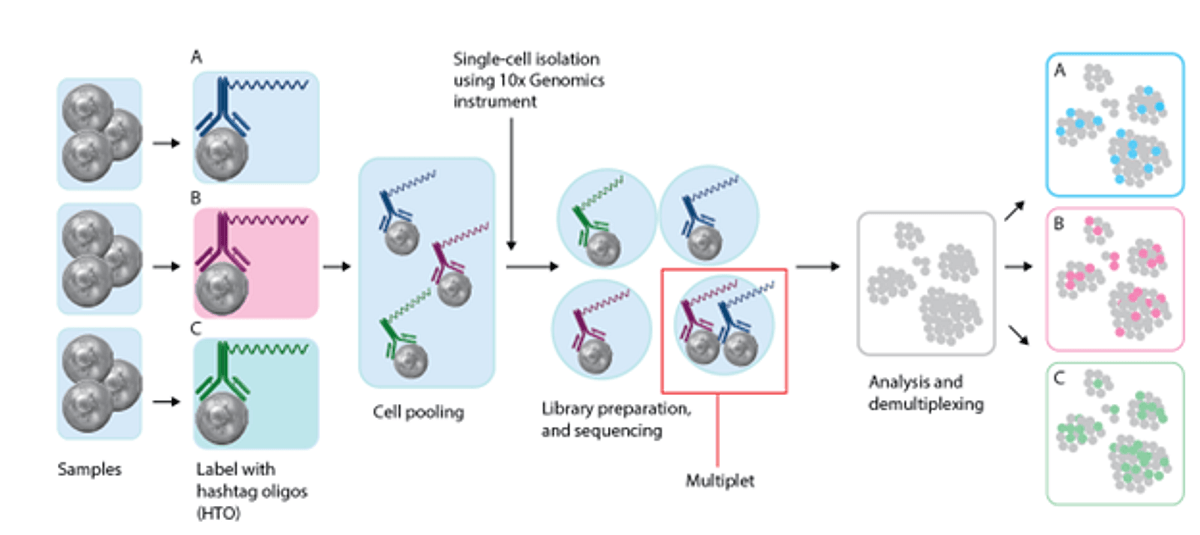
Figure 1: Multiplexing workflow using TotalSeq hashtags.
Human PBMCs were isolated from healthy donors. Hashtag antibodies (500 ng total for TotalSeq-A and 50 ng for TotalSeq-B and TotalSeq-C) and the corresponding TotalSeq Human TBNK Panel cocktail were mixed at the recommended concentration to stain PBMC.
The 10x Genomics Single Cell 3’ Reagent Kit v3.1 (used with TotalSeq-A and -B) and the Chromium Next GEM Single Cell V(D)J Reagent Kits v1.1 (used with TotalSeq-C) were used for scRNA isolation. Please note that this data set was generated using 10x Genomics single index libraries. TotalSeq A libraries were generated according to manufacturers instructions and Bio Legend’s TSA single index protocol. In consideration that 10x has discontinued the single index assays, BioLegend now offers an alternative dual index TSA protocol and TotalSeq-B or -C for use with 10x Genomics dual indexing kits that users can use moving forward.
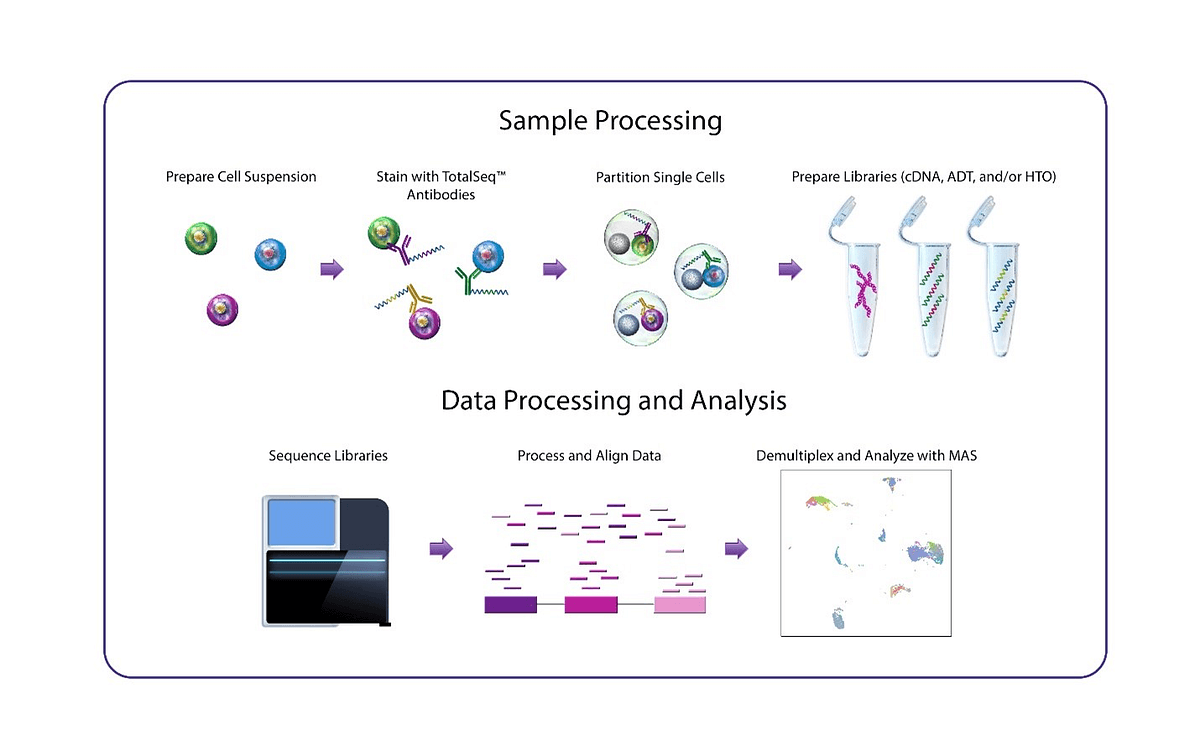
Figure 2: Generation of cDNA, ADT and HTO BioLegend libraries, cDNA, ADT and HTO libraries were combined and sequenced on the NextSeq 2000 and NovaSeq 6000 with the following run configuration: 28 + 8 + 91 and 1% PhiX spike in. The resulting FASTQ were analyzed with DRAGEN single cell RNA (see our coming blogpost on that topic).
NextSeq 2000 run: https://basespace.illumina.com/s/JDQaQ1KPgMmT
NovaSeq 6000 run: https://basespace.illumina.com/s/cSSGOzasyYFE
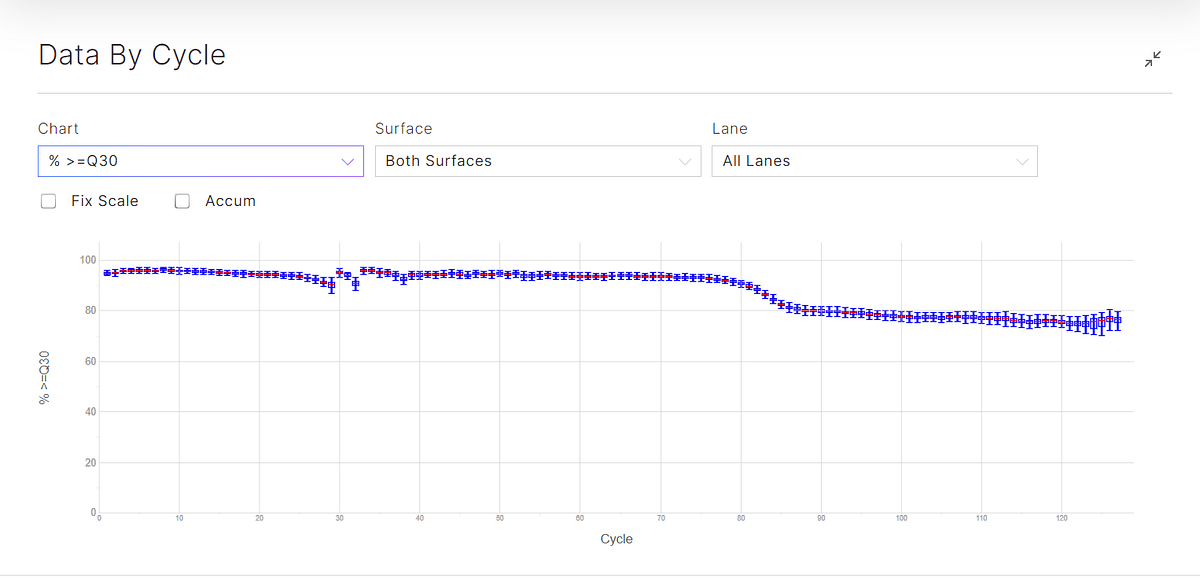
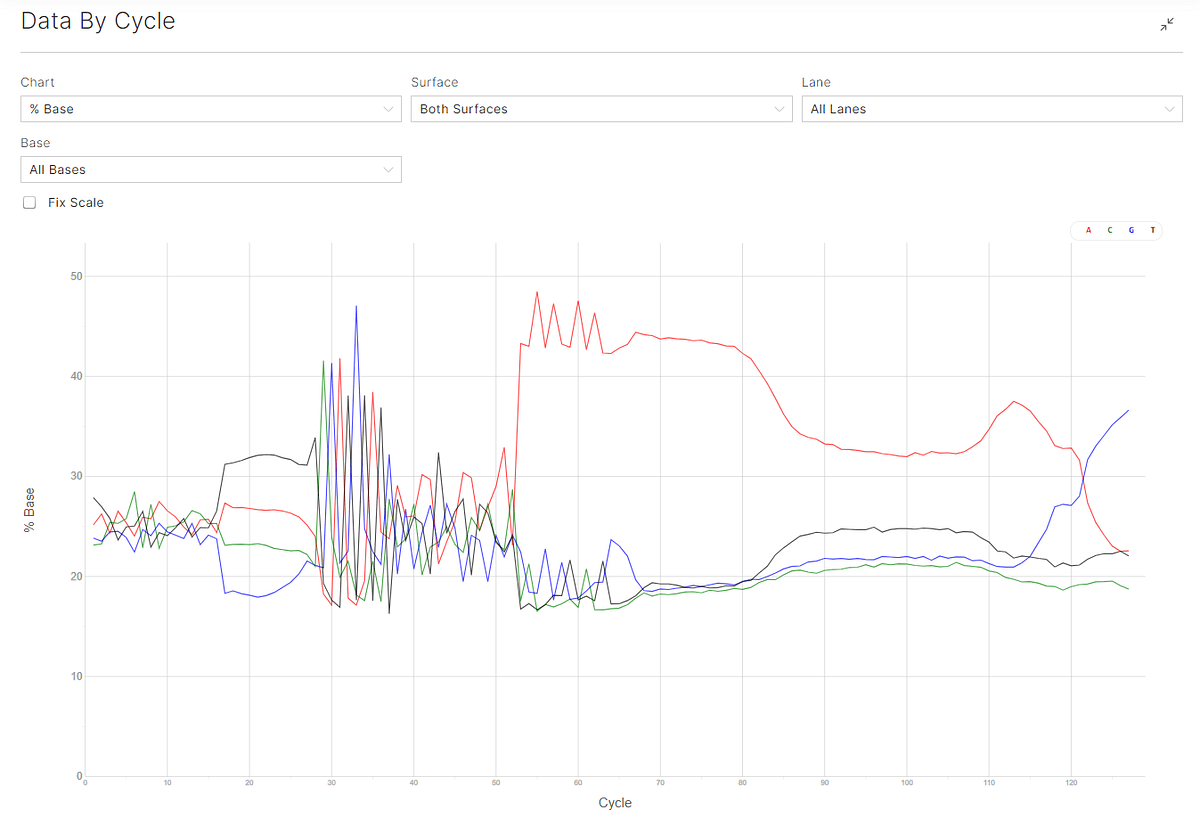
Figure 3: Q30 plots (top) and % base plots (bottom) for the sequencing runs. Both ADT and HTO libraries have very short inserts, especially in the reverse reads, resulting in an A-overcall and expected Q30 drop after 55 cycles of sequencing.
Additional resources:
- https://www.biolegend.com/en-us/totalseq?gclid=Cj0KCQjwnNyUBhCZARIsAI9AYlEQLRfJqipvEJw41tStTZNt1ra9HOra4AOzU3AZCX9gjl1owrlqHnUaArnSEALw_wcB
- https://www.biolegend.com/en-us/cell-hashing-app-note
- https://www.biolegend.com/en-us/protocols/totalseq-a-antibodies-and-cell-hashing-with-10x-single-cell-3-reagent-kit-v3-3-1-protocol
- https://support.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/biolegend-ben-seq-app-note-m-gl-00024/biolegend-ben-seq-app-note-m-gl-00024.pdf
- https://www.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/biolegend-totalseq-dilution-application-note-m-gl-00475/biolegend-totalseq-dilution-application-note-m-gl-00475.pdf
Support
Contact BioLegend for any questions regarding the antibodies, the library prep assay and the downstream analysis. Contact Illumina Tech support for sequencing related questions.
Special thanks to our colleagues at BioLegend for providing all the libraries, and to our colleague Robin Bomnbardi from the Emerging Apps team to sequence them.
For Research Use Only. Not for use in diagnostic procedures. M-GL-01268