- Home
- News & Updates
- DRAGEN: Simple Integration | Enhanced Analysis
-
DRAGEN
-
News
- 05/20/2020
DRAGEN: Simple Integration | Enhanced Analysis

With advances in next-generation sequencing (NGS) technology, the volume of generated sequencing data continues to grow exponentially. This growth requires fast and efficient analytical methods that maintain high standards in accuracy for variant calling. Customers may struggle with updating their existing bioinformatics infrastructure to keep up with the vast amount of data and increases in computing power necessary for analysis of new applications that require deeper sequencing. To address these challenges, Illumina offers the DRAGEN (Dynamic Read Analysis for GENomics) Bio-IT Platform. DRAGEN can be easily integrated into existing high-performance computing (HPC) or cloud-based solutions to enhance operations. This blog will walk through some examples of integrating DRAGEN within typical scenarios.
Why labs integrate DRAGEN:
- Increased operational efficiency: DRAGEN can process a whole genome in under 25 minutes and an exome in < 8 minutes, reducing turnaround times and enabling labs to process more data in less time.
- Scalable operations: The speed and versatility of DRAGEN enables labs to expand their bandwidth and capabilities without needing to create pipelines and invest in new HPC clusters; a single DRAGEN Server can run all DRAGEN pipelines, supporting a vast array of experiment types.
- Reduced maintenance: DRAGEN customers can take advantage of quarterly software updates, including new features, improvements, and functionality.
- Considerable cost savings: DRAGEN offers a smaller physical footprint, reduced hardware costs, and diminished power and cooling requirements.
Common IT infrastructures
DRAGEN Servers can be implemented in a central, regional, or cloud- based infrastructure. We'll explore the most common implementation below.
Centralized DRAGEN on-site Deployment
Common academic infrastructures typically consist of individual labs, each with their own fleet of sequencers, temporarily storing raw sequencer output data, ie, BCL or cBCL files, on cache storage, such as a Network Attached Storage (NAS), proximal to their instruments (Figure 1). The DRAGEN Server nodes are centrally managed as part of an institutional compute cluster and analyses are queued and managed through a workload manager such as SLURM.
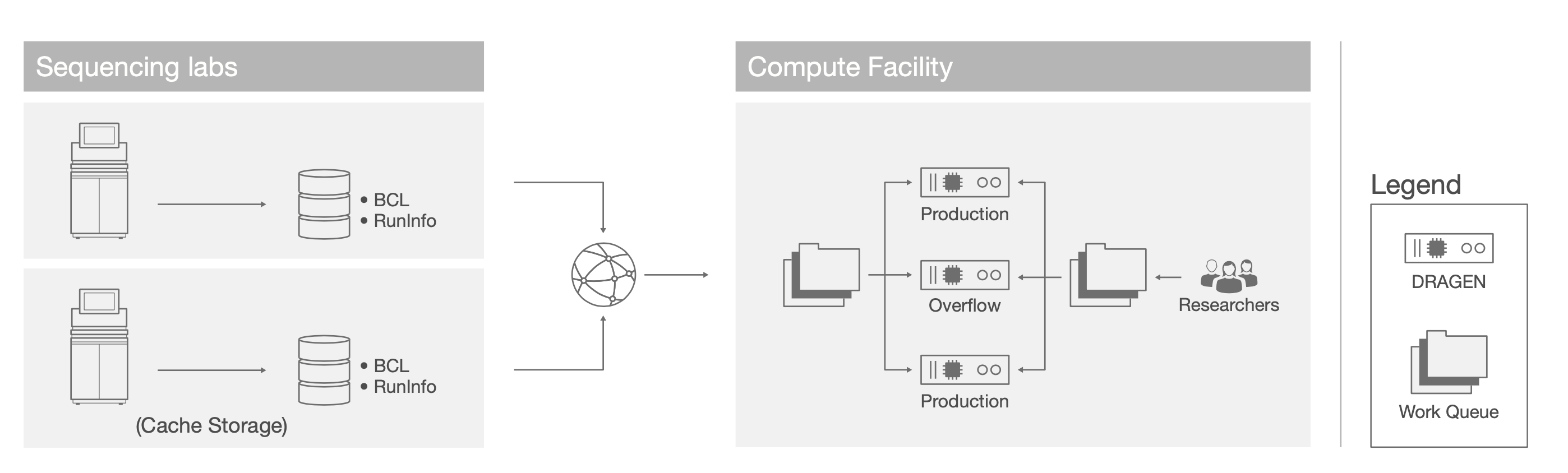
Regional DRAGEN On-site Deployment
Common core lab or translational lab infrastructures consist of each sequencing lab purchasing and using their own fleet of DRAGEN servers for rapidly processing sequencing data from BCL or cBCL files to VCF or gVCF files (Figure 2). Sequencing core labs can use the DRAGEN Platform to perform data quality control (FastQC) and read trimming (poly-G trimming or Trimmomatic) themselves, without transferring FASTQ data to a bioinformatics team or a compute facility. This reduces inefficient back-and-forth communication between teams due to flagged QC failures.
These analyzed data can then be uploaded to a central compute facility for downstream analysis, variant annotation, data sharing, and alignment against other genome references.
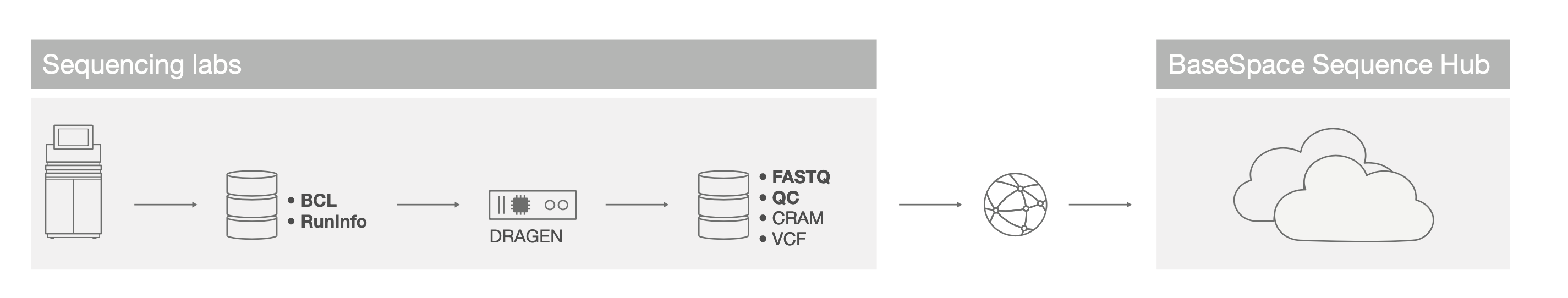
Figure 2: Regional DRAGEN on-site deployment—Core labs can locate DRAGEN Servers within the sequencing facility to process data rapidly and minimize network file transfers.
Hybrid Cloud Infrastructures
Some customers currently split analysis and storage functions between on-premise and cloud-based functions (Figure 3). Using Application Programming Interface (API) connectivity between the DRAGEN Server and the Illumina Analytics Platform (IAP, available Q3/Q4 2020), customers can customize pipelines in the cloud for sharing across networks. As in the regional DRAGEN deployment scenario, secondary analysis is accomplished with on-premise DRAGEN servers, and the resulting data are transferred to another resource for further processing. In a hybrid cloud scenario, this usually involves a platform-as-a-service provider such as BaseSpaceTM Sequence Hub. Cloud services offer a richer experience for data management, such as easy data archiving and integration with third- party partners and vendors for downstream tertiary analysis.
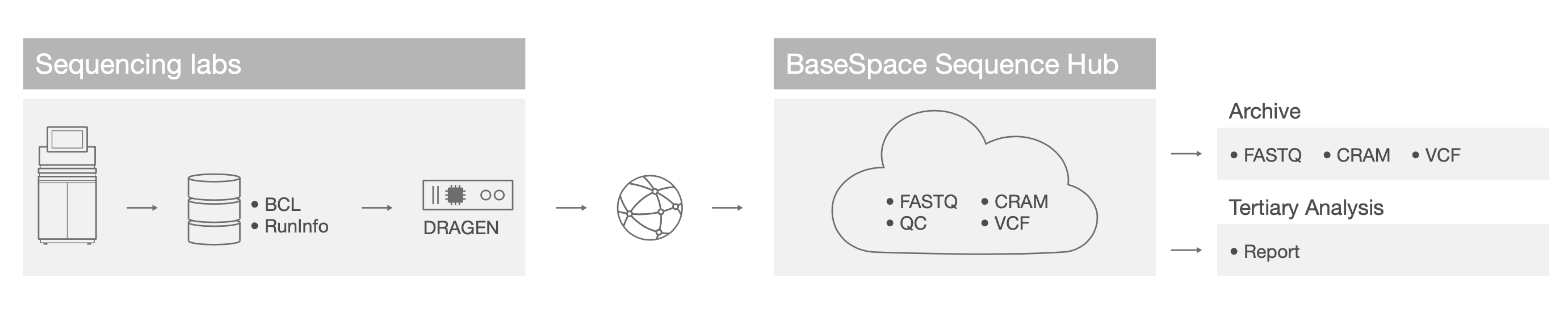
Figure 3: Hybrid cloud DRAGEN deployment—Customers can use a DRAGEN Server to perform primary and secondary analysis, and harness cloud resources, such as BaseSpace Sequence Hub, for multi-sample analysis, data archive, and tertiary analysis.
Cloud deployment
Customers can also forego installation and management of a DRAGEN Server and choose to leverage DRAGEN Applications available in the cloud via BaseSpace Sequence Hub or Amazon Web Services (AWS) Marketplace for primary and secondary data analysis (Figure 4).
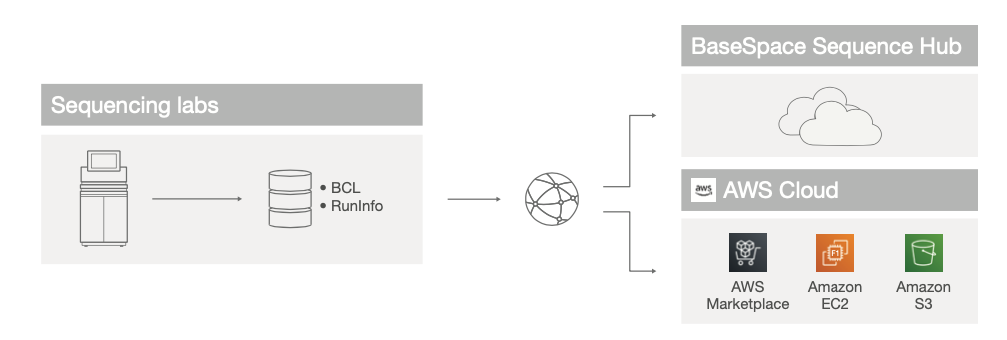
Figure 4: Cloud DRAGEN deployment—Customers can use DRAGEN Applications available in the cloud via BaseSpace Sequence Hub or AWS.
For research use only. QB#9656