- Home
- News & Updates
- DRAGEN Two-Year Update
-
DRAGEN
-
News
- 05/15/2020
DRAGEN Two-Year Update
Today marks the two-year anniversary of Illumina’s acquisition of Edico Genome and the DRAGEN™ bioinformatics technology. The past two years have been filled with new developments, partnerships and advances all centered around a singular key theme – our continued commitment to deliver more value to our customers.
With the integration of DRAGEN into the Illumina product family, we’ve been able to rapidly grow DRAGEN’s functionality and accuracy thanks to the newfound access to some of the best researchers, scientists and growing install base. Two-years in, we’re excited to reflect back on our growth and explore how we’re enabling researchers to ask new questions and glean more valuable data with our expanded portfolio.
Delivering more valuable data
We’re committed to delivering more accurate data and deeper insights to enable our customers to unlock new discoveries.
To that end, the DRAGEN development team has been hyper-focused on increasing the value of data analyzed via DRAGEN through continuous accuracy improvements, inclusion of new variant callers and tools, and the integration of new metrics and reports. As part of Illumina, we’ve been able to integrate the best of Illumina’s legacy tools and methods into DRAGEN and have the distinct pleasure of collaborating with top bioinformaticians around the globe to continuously tweak our methods to deliver more accurate solutions. I’ll dive into a few focal areas below.
Somatic Accuracy Improvements
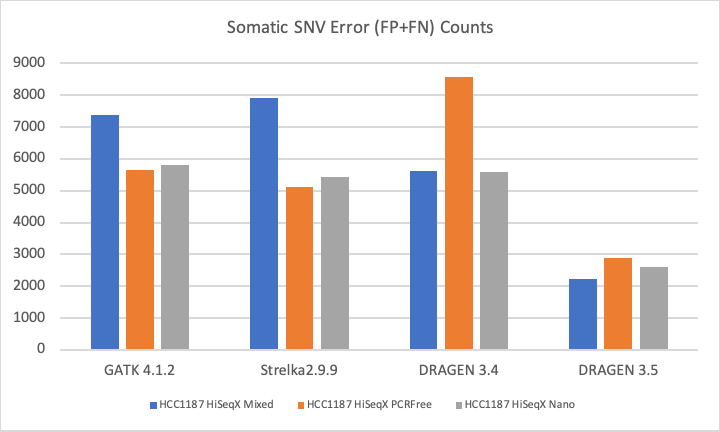
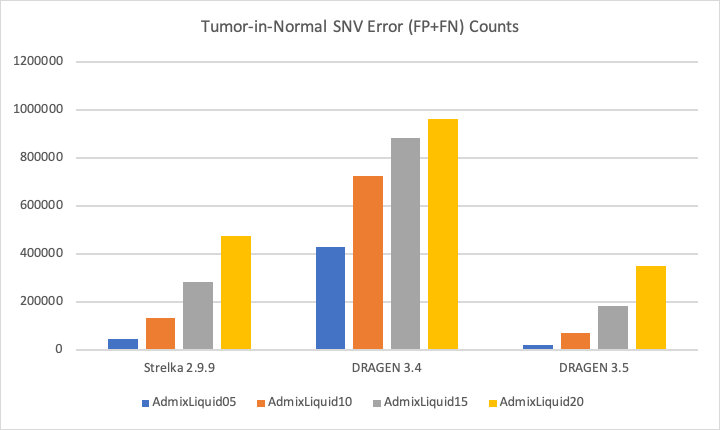
The DRAGEN Somatic Pipeline was a large focus area in 2019. The latest DRAGEN release, v3.5, delivered significant boosts in both sensitivity and precision, demonstrated in the figure below. In addition to accuracy gains for solid tumors, DRAGEN now supports hematological cancers, and is capable of handling higher TiN (tumor in normal) level and lower purity.
RNA Fusion Detection
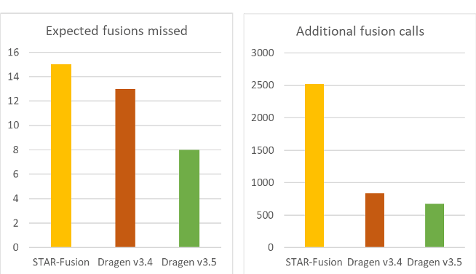
DRAGEN v3.5 delivered significant improvements in sensitivity and speed for RNA fusions detection. To address the challenges with RNA-Seq fusion detection, we created a repository of 153 samples with known validated or expected fusions from 5 different sources representative of real-world applications. The sources included libraries from whole RNA and enrichment cell lines, fresh leukemia, and FFPE sarcoma, resulting in 223 expected fusions from 153 samples.
The resulting gains can be seen in figure 1 below.
It’s not all about accuracy and new features. Last Fall we announced a new partnership with the Broad Institute to transfer DRAGEN’s Germline Small Variant Caller to the Broad for integration in the soon-to-be-released DRAGEN-GATK Best Practices. As part of our partnership, we’ll be offering our non-accelerated DRAGEN Mapper and DRAGEN Germline Small Variant Caller to the community on GitHub. Part of the beauty of our partnership is the agreed upon functional equivalency between the open sourced DRAGEN-GATK Best Practices and Illumina’s accelerated DRAGEN Germline offerings. Why is this important? With the release of DRAGEN-GATK Best Practices, researchers will be able to seamlessly compare and collaborate with data generated with open sourced and Illumina-offered DRAGEN tools, reducing the risk for batch effect.
While accuracy and compatibility are both essential, the true value of genomic data comes from being able to put the data into context to answer important questions and facilitate new discoveries. We’ve integrated DRAGEN into a number of end-to-end, application-specific workflows to enable researchers to seamlessly push DRAGEN outputs into interpretational and reporting tools.
Making it simpler to find answers
Another key focal area was simplicity through integration. As part of the Illumina portfolio, DRAGEN has been integrated with Illumina’s sequencers, library prep kits and its comprehensive software suite, making it easier for researchers to implement and scale workflows. The end goal of this effort is to enable researchers to focus on the science and discoveries, not the process.
We’ve done this in a few key areas:
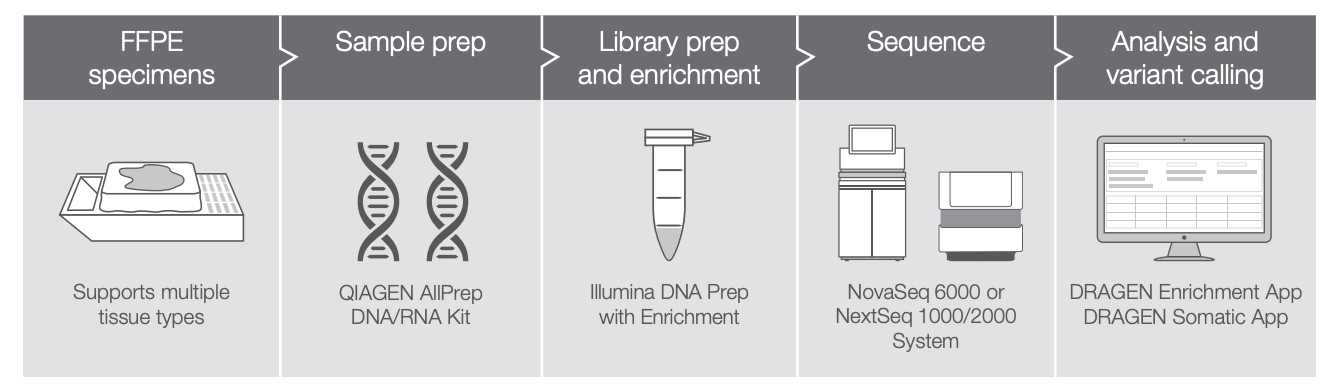
+ Enabling end-to-end applications: DRAGEN is now embedded in many of Illumina’s application-specific end-to-end workflows, which are designed to help regulated customers navigate from sample to answer. Using the example of Illumina’s TruSight Oncology 500 ctDNA, the integration of DRAGEN drastically reduces turnaround time from ~nine days down to 20 hours while ensuring high data accuracy and reliability. Through these end-to-end solutions, researchers can seamlessly push DRAGEN outputs into downstream interpretation tools to find the answers they seek.
+ Deeper integration with Illumina’s portfolio: DRAGEN has become a core component of Illumina’s product development, translating to new features and pipelines that are optimized to be paired with Illumina’s upstream software and library preparation kits. We’ve also integrated the full suite of DRAGEN Pipelines on BaseSpace Sequence Hub, enabling researchers to seamlessly stream data to the cloud for push-button analysis via DRAGEN at a low cost-per-sample. And, of course, we’ve integrated DRAGEN into the new NextSeq™ systems to streamline and accelerate downstream analysis.
+ More comprehensive pipelines: One of the first major DRAGEN releases at Illumina featured an easier-to-use, comprehensive DRAGEN Germline Pipeline that brought CNV (copy number variant), SV (structural variant), and Repeat Expansion detection into a singular pipeline. Since then, we’ve continued to expand and strengthen our pipelines with the addition of new metrics, variant callers and tools to deliver more comprehensive solutions to our customers.
+ Easy-to-use Apps with Built in Reporting: For researchers desiring a simplified approach to data analysis, we now offer our full suite of DRAGEN Pipelines on BaseSpace Sequence Hub, many of which include built-in data visualization and reporting functionality. The DRAGEN apps in BaseSpace offer a simple, low cost-per-sample. The DRAGEN Enrichment App is a great example of this.
Greater Accessibility
Every lab has a preference for their computing environment, and whether that’s a customized cloud solution or a local on-premises setup, we’ve there to support our customers in the environment of their choosing. When DRAGEN first came to market back in 2015, it was accessible on-premises via a local DRAGEN Server. Today, we have five access points, enabling our customers to harness the power of DRAGEN where they want it, when they want it. And with the completion of DRAGEN-GATK later this year, users will also be able to harness the open sourced, non-accelerated DRAGEN Germline Pipeline for small variant calling on GitHub, rounding us up to a nice six.
To recap, we’ve included a quick list of available access points below.
- DRAGEN Server: our DRAGEN Server has now spread to all corners of the globe, and with the new DRAGEN v3 Server released earlier this year, the DRAGEN Server continues to be our fastest access point.
- DRAGEN on BaseSpace Sequence Hub: for our customers seeking a simple, affordable push-button solution, we now offer all DRAGEN Pipelines on BaseSpace Sequence Hub at a low cost per sample (average ~$5/genome).
- DRAGEN on NextSeq 2000: Perhaps one of the most exciting advancements over the past two years has been the integration of DRAGEN on the new NextSeq systems, allowing for efficient, accurate analysis at the edge, now all-in with sequencing.
- DRAGEN via AWS Marketplace: For customers desiring to run DRAGEN within their own compute environment, DRAGEN can be run via an AMI (Amazon Machine Image) on AWS Marketplace.
- DRAGEN API: The newest member of the DRAGEN environment family is the DRAGEN API. Available in early access, advanced clouds users desiring automation and a high degree of flexibility can leverage DRAGEN via an API.
It has been a great first two years, and we’re looking forward to many more to come. We’d like to thank all of our customers, partners and collaborators that have contributed to the growth and progression of DRAGEN throughout the years.
For research use only.