- Home
- News & Updates
- DRAGEN v3.6: up to 40% faster
-
DRAGEN
-
News
- 07/17/2020
DRAGEN v3.6: up to 40% faster
DRAGEN v3.6 webinar: Principal Software Engineer Shyamal Mehtalia shares a robust overview of new features, speed and accuracy gains
We often talk about DRAGEN as a combination of hardware and software that work together to help our customers get the most out of their data and computational resources. DRAGEN utilizes all of the resources available to it, including CPUs, memory, and Non-Volatile Memory Express (NVMe) SSD storage, but the lion’s share of the speed comes from offloading computationally expensive portions of analysis pipelines to the FPGA card.
Doing so is not straight forward, however, as “programming” an FPGA is actually done by designing custom low-level hardware circuits that must work in coordination with the main system components. Hardware design at this level requires expert knowledge not widely distributed among the developer community. Even when you are able to implement something within a FPGA, the implementation may not be efficient enough to overcome the overhead from moving data to and from the FPGA card memory.
With that background, we are excited to announce that the DRAGEN team has implemented deBruijn graph constructions to the FGPA. This means that for our cloud and DRAGEN V3 Servers, the germline and somatic small variant calling pipelines are up to 40% faster!
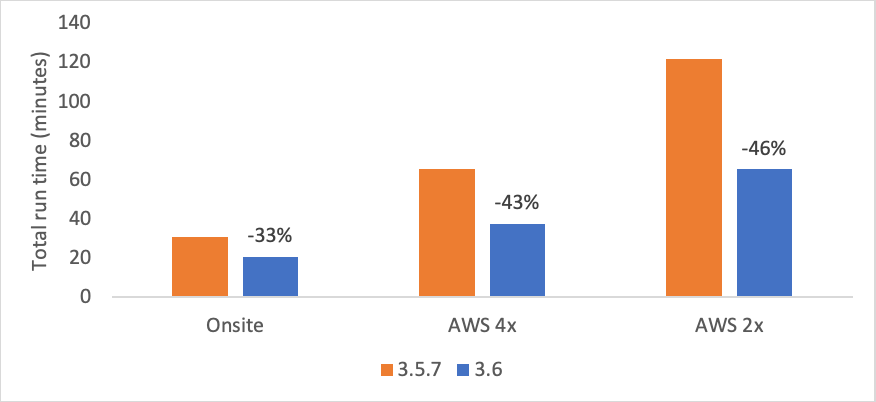
Significant improvements in run times compared to v3.5 achieved through the addition of hardware accelerated graph building. Whole genome sequencing at 38x coverage (123 Gb) with VCF and BAM generation.
As you can see from above, the runtimes are greatly reduced, with the largest gains occurring on the Amazon Web Services (AWS) F1 instances, which underpin our BaseSpace DRAGEN applications. These faster runtimes on BaseSpace will result in lower price per analysis. There is good news for DRAGEN Server V3 customers as well, as they will be able to analyze more data with their existing hardware.
Another significant update to DRAGEN v3.6 is the addition of FastQC-like metrics to the pipeline output. The FastQC metrics are calculated on-the-fly as reads are mapped to a reference within the FPGA. This means that FastQC metrics are now a core part of DRAGEN and users can get this output ~7x faster than using the open source CPU-only version of FastQC. You can visualize the output DRAGEN FastQC statistics using MultiQC, which will become integrated with most DRAGEN BaseSpace Sequence Hub Apps moving forward.
Other DRAGEN additions and improvements for v3.6 include:
- Poly-G trimming is now implemented in the FPGA, allowing for real-time soft-trimming of reads before they are aligned to a reference by the DRAGEN mapper. Poly-G artifacts appear on 2-channel sequencers when the “dark” base G is called after synthesis has terminated. In the new trimmer, the G’s from the incoming read are not stripped and are still in the output, leading to improved mapping accuracy without any loss of original information. There is an option for hard-trimming, which does permanently trim the poly-G from the incoming read. In both cases, statistics are output from the trimmer of the poly-G. Note that this feature is also only available for customers with DRAGEN V3 servers or on the cloud.
- The PopGen cohort analysis tools see several improvements, including 3X faster runtime for the JointGenotyper, and adding the capability of the GvcfGenotyper to import both GATK gVCFs and multi-sample pedigree gVCFs.
- Improving the somatic CNV caller for better handling of noisy-depth segments, such as those that appear in FFPE samples. We also improved the exome CNV caller to handle errors from panel of normal samples.
- Small variant calling improvements include gains in indel precision without sacrificing sensitivity, as well as the ability to designate specific genomic loci as somatic “hotspots” where the risk for genetic mutations is significantly elevated, thus increasing sensitivity in those regions.
- The methylation pipeline now supports TAPS (TET-assisted pyridine borane sequencing), which is a new assay that directly converts methylated C to T, enabling lower input DNA and better preserving genomic complexity compared to traditional methods.
And much more! As always you can get more details in the DRAGEN Release Notes page on our support site here.
DRAGEN v3.6 Hardware Support
Moving computational operations from the CPU to the FPGA does not come for free. Namely with each additional function, the FPGA has to have the physical resources to support the data structures and computational circuits to support the function. Unfortunately, older DRAGEN servers do not have the necessary FPGA hardware that made offloading of the deBruijn graph possible. Only DRAGEN servers with a Xilinx U200 FPGA can support the graph offload for faster runtime, as well as the FastQC and Poly-G trimming. Customers with older hardware will still benefit from our additional features and improved accuracy coming in DRAGEN v3.6 software.