- Home
- News & Updates
- DRAGEN v3.7: Single Cell RNA, PrecisionFDA Accuracy Gains, and More
-
DRAGEN
-
News
- 11/10/2020
DRAGEN v3.7: Single Cell RNA, PrecisionFDA Accuracy Gains, and More
In addition to being laser focused on improving DRAGEN’s accuracy and speed, we also strive to continually expand DRAGEN capabilities. When it was first introduced, DRAGEN provided rapid and accurate germline small variant calling. Over the years, we’ve added additional variant types, expanded our offering to cover tumor and tumor/normal, introduced RNA and methylation pipelines, and upped our game in offering targeted callers. You could say we’re on a mission to deliver the maximum information from sequencing.
With the release of DRAGEN v3.7, we take a major step forward by expanding our capabilities to include a Single Cell RNA pipeline, full UMI support, and a targeted CYP2D6 caller.
In addition to all of the new features with this release, we’ve also introduced substantial accuracy gains in both germline and somatic pipelines. For germline small variant calling, you’ll see the innovations that powered our prize-winning performance in the PrecisionFDA Truth Challenge V2 in this release, including a graph capable reference genome. We also provide significant accuracy improvements in our somatic pipeline, including a 5x reduction in false positives compared to DRAGEN v3.6 in tumor only mode.
Buckle up for a whirlwind overview of a few of the major highlights from DRAGEN v3.7.
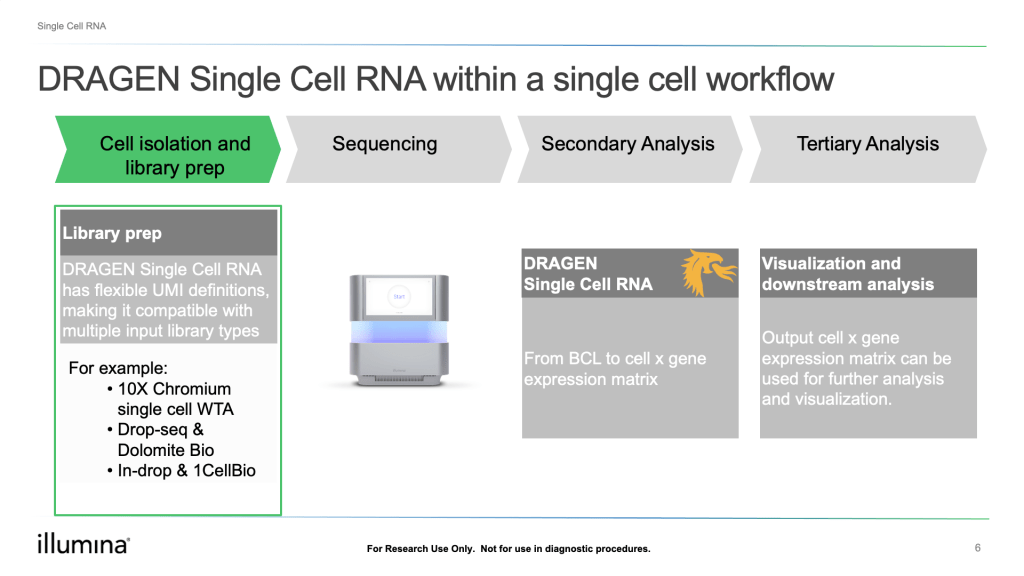
Single Cell RNA
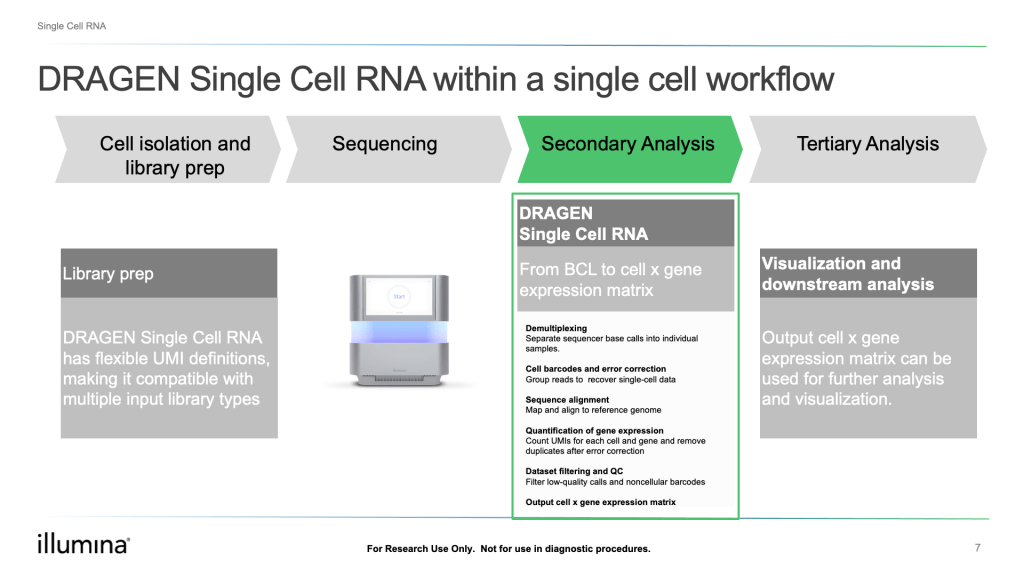
DRAGEN’s efficiency and speed can now be leveraged to accelerate single cell gene expression experiments. DRAGEN Single Cell RNA is an integrated pipeline that goes from base calls to cell by gene expression matrix with a single user touchpoint. The pipeline is available for on-premise DRAGEN servers, via AWS Marketplace, as well as onboard the NextSeq 1000/2000 (Illumina’s latest sequencer that has DRAGEN built into the instrument). If you’re running the pipeline onboard the NextSeq 1000/2000, you’ll have the added benefit of being able to queue up analysis when you plan your sequencing run, which means you’ll have a cell by gene expression matrix and quality control metrics when your run is done in addition to BCL and FASTQ files. The output cell x gene expression matrix is compatible with popular single-cell analysis tools such as Scanpy, AnnData, and Seurat.
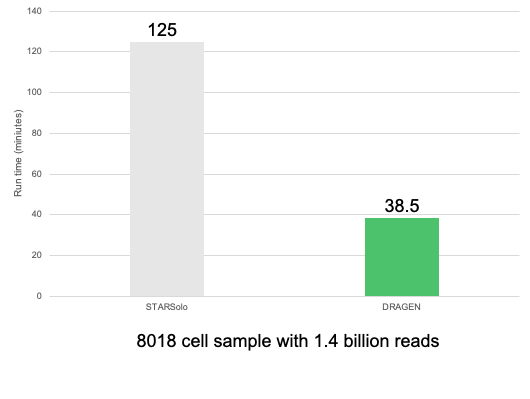
The pipeline has high performance and ultra-fast run times due to the DRAGEN FPGA accelerated mapper and efficient engineering; only two passes over the data are performed throughout the pipeline with minimal disk I/O. That means it’s 3x to 25x faster than any comparable open source or commercially available tool we (or early access customers) have tested it against.
In addition to speed, there are two additional benefits that set DRAGEN Single Cell RNA apart:
- Flexible UMI definitions make this pipeline compatible with multiple input single cell expression library prep types. Different lengths and locations for both cell barcodes and UMI are supported, enabling error correction for random or non-random barcode designs and UMI error correction.
- You get full RNA alignments for transcriptome reads. Unlike kmer-based methods that do not provide alignments and only consider the known transcript sequences, DRAGEN performs alignments to a complete genome (including novel isoforms and intergenic reads).
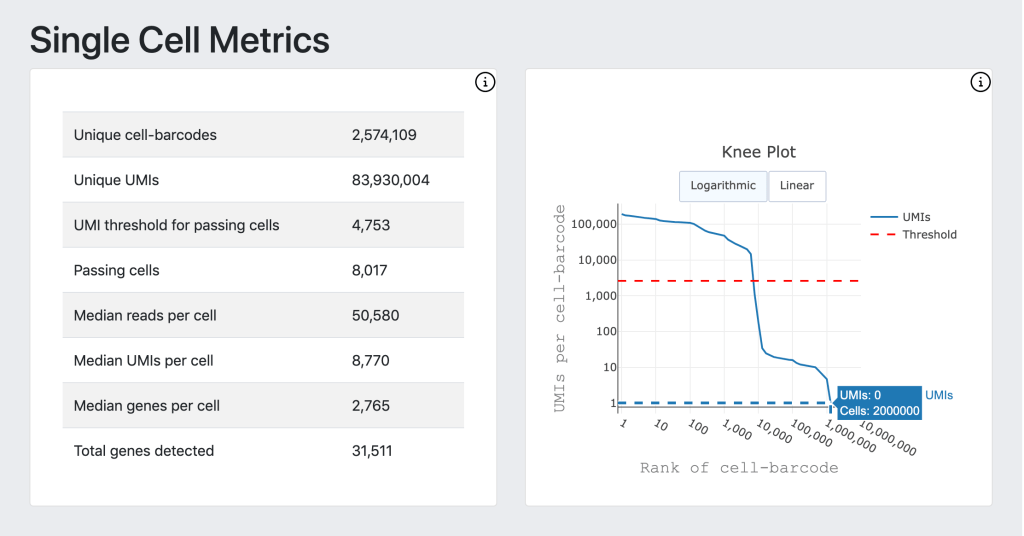
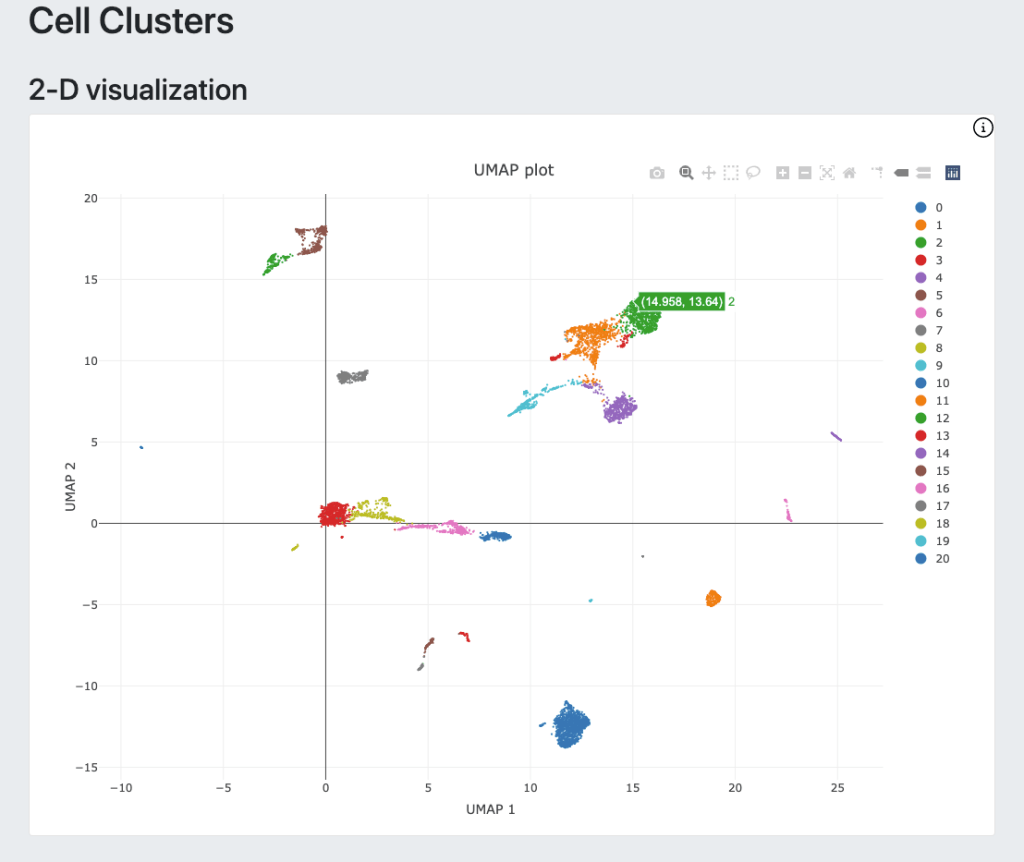
DRAGEN Single Cell RNA is highly concordant with established tools. When we compared to STARsolo, we observed high correlation of per-cell gene expression and very similar Scanpy ‘Leiden’ clustering on the output cell x gene expression matrix.
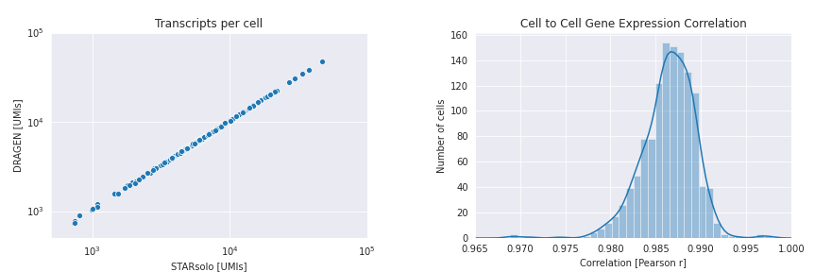
857 / 858 cells overlap
STARSolo gitub: https://github.com/alexdobin/STAR/blob/master/docs/STARsolo.md
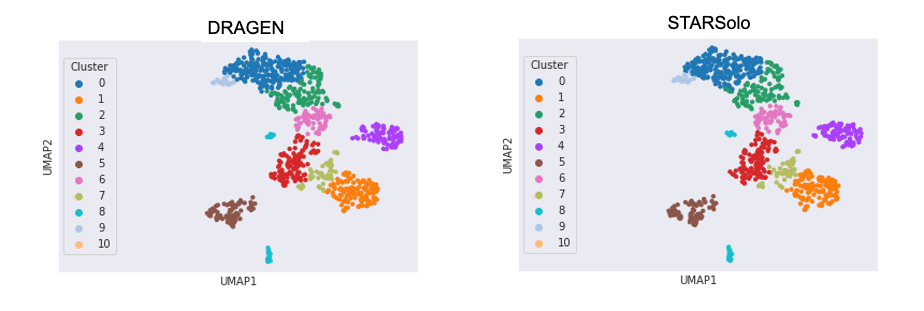
853 out of the 857 cells are assigned to the same cluster.
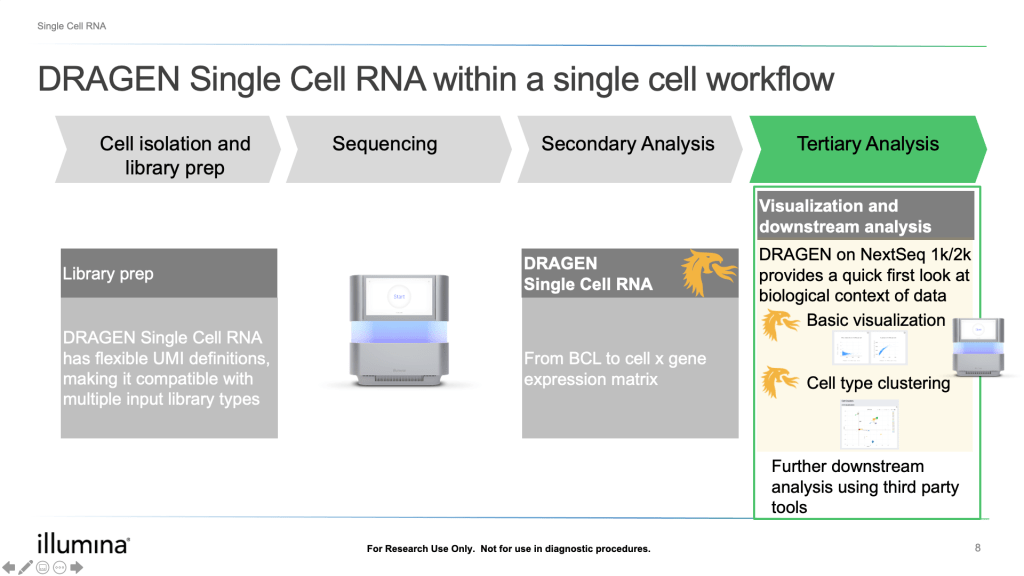
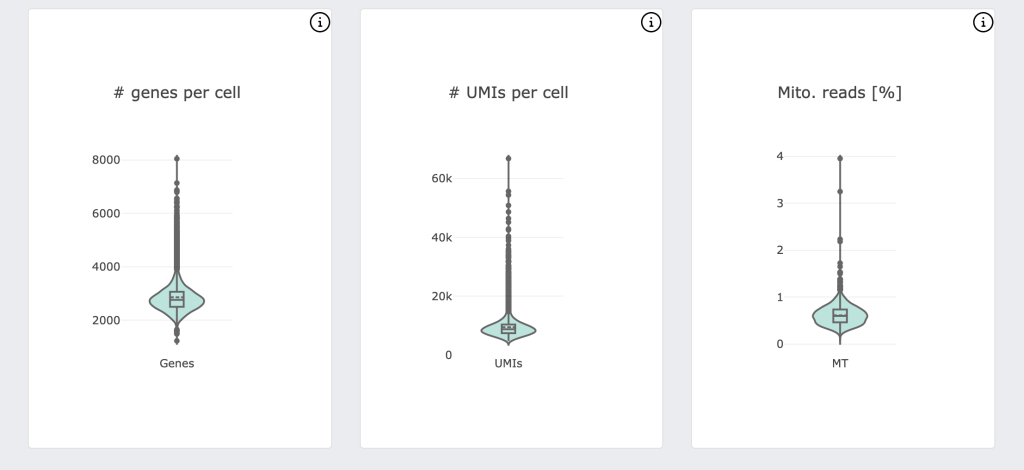
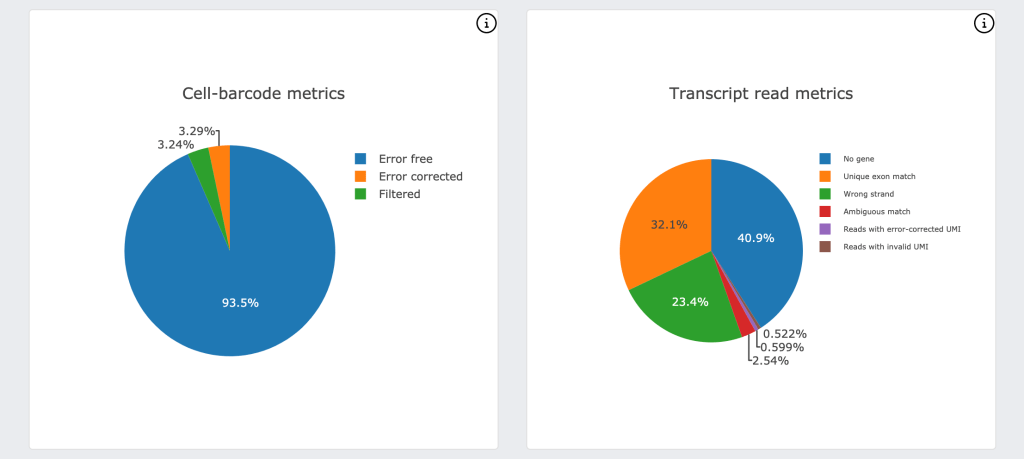
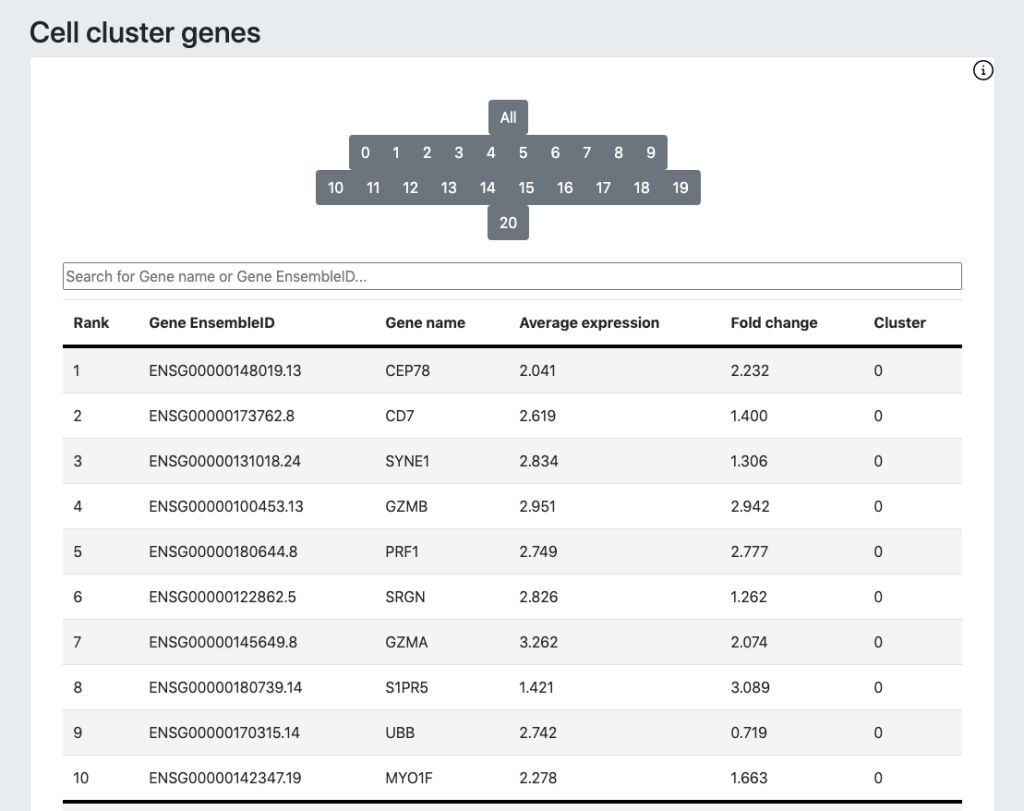
For DRAGEN on NextSeq 1000/2000, the Single Cell RNA pipeline will also output a downstream html report that provides a quick first look at the biological context and quality of your data, including a UMAP projection, automated graph-based clustering (SAM + Leiden clustering) and cluster marker gene detection.
We hope that leveraging the speed of DRAGEN for your single cell expression experiments will help accelerate the pace of your research.
Full UMI support
With DRAGEN v3.7, we now offer support for all UMI types: random-simplex (Agilent HS1), random-duplex (IDT xGen Duplex Seq), and non-random duplex (TruSight UMI). We also introduce a new probabilistic UMI collapsing model with improved accuracy. Our UMI pipeline is 15-25x faster than other methods (Fgbio) and has ~30% lower error rate.
We’re also introducing UMI-aware variant calling for targeted panels, which prevents read over-counting, boosts sensitivity for driver mutations, and allows for a customized panel of normals, ultimately leading to greater sensitivity and reduced errors. This is particularly useful for liquid biopsy samples.
CYP2D6 Caller
CYP2D6 genotyping is now integrated with DRAGEN for use with germline WGS data. The genotyper is based on the open-source tool Cyrius and yields 100% concordant results in a 143 sample data set. It outputs the star allele diplotype for each sample, and because over 120 CYP2D6 star alleles are supported, DRAGEN can call a definitive genotype in nearly all samples. The CYP2D6 caller can be enabled to run in parallel with other components in a germline WGS analysis.
Germline accuracy gains (PrecisionFDA)
We’re excited to release two major innovations the team worked on leading up to the PrecisionFDA challenge that enabled us to perform so well in that challenge: graph capable mapping and joint detection of overlapping variants.
The graph capable mapper in DRAGEN is a key enabler in improving variant calling accuracy in segmental duplications and other regions previously difficult to map with Illumina reads. We’ve also introduced joint detection of overlapping variants, which gives a significant accuracy boost in regions where SNP and INDELs overlap or are near short tandem repeats.

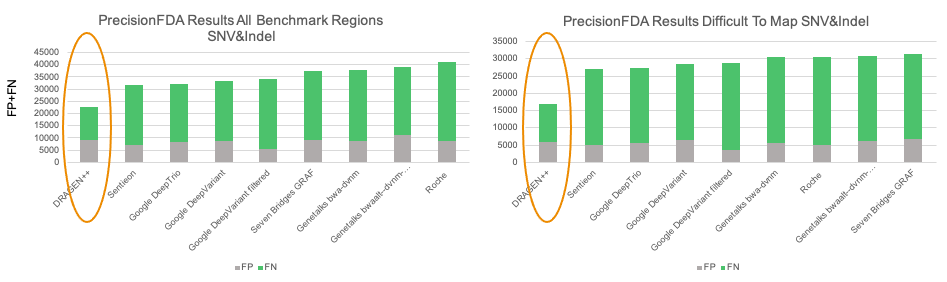
For a deeper dive into the innovations powering our PrecisionFDA wins, check out this article on our Research and Innovation page.
Somatic accuracy gains
We’ve also introduced significant accuracy improvements for tumor only analysis, and now boast 5x fewer false positives compared to DRAGEN v3.6 and sensitivity >95%. We’ve updated the genotyping approach for tumor only mode, which is now the same as tumor/normal mode and leads to overall improved precision.
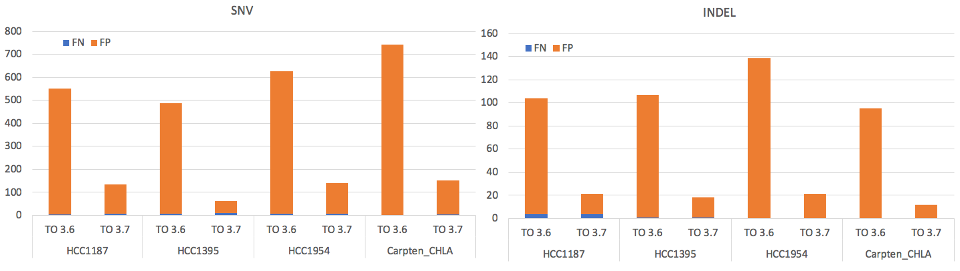
We now have improved accuracy in tumor only mode for WGS, WES, and Panels. You’ll also see accuracy improvements in WGS and WES tumor/normal mode.
This release was packed full of new functionality and accuracy improvements, and we’ve only touched on the tip of the iceberg here. For a more comprehensive overview, check out the release notes and the webinar.