- Home
- News & Updates
- DRAGEN v4.3 – Powering a comprehensive genome with industry-leading innovations
-
DRAGEN
-
Product updates
-
News
- 06/11/2024
DRAGEN v4.3 – Powering a comprehensive genome with industry-leading innovations
Introducing DRAGEN v4.3! This latest version brings a full suite of innovative features enabling unparalleled genomic analysis.
DRAGEN v4.3 comes with a next-generation multigenome (graph) mapper that now incorporates 128 high quality assemblies from internally built pangenome data. It also has a new machine learning mosaic model in the small variant caller, and a new family of specialized callers such as Multi-Region Joint Detection (MRJD) that can be used for screening in segmental duplication regions for mutations in difficult genes such as PMS2 for hereditary cancer.
Access the latest software here and learn more about new features on our new gitbook page.
Let’s take a closer look at some of the major improvements.
Next generation multigenome technology
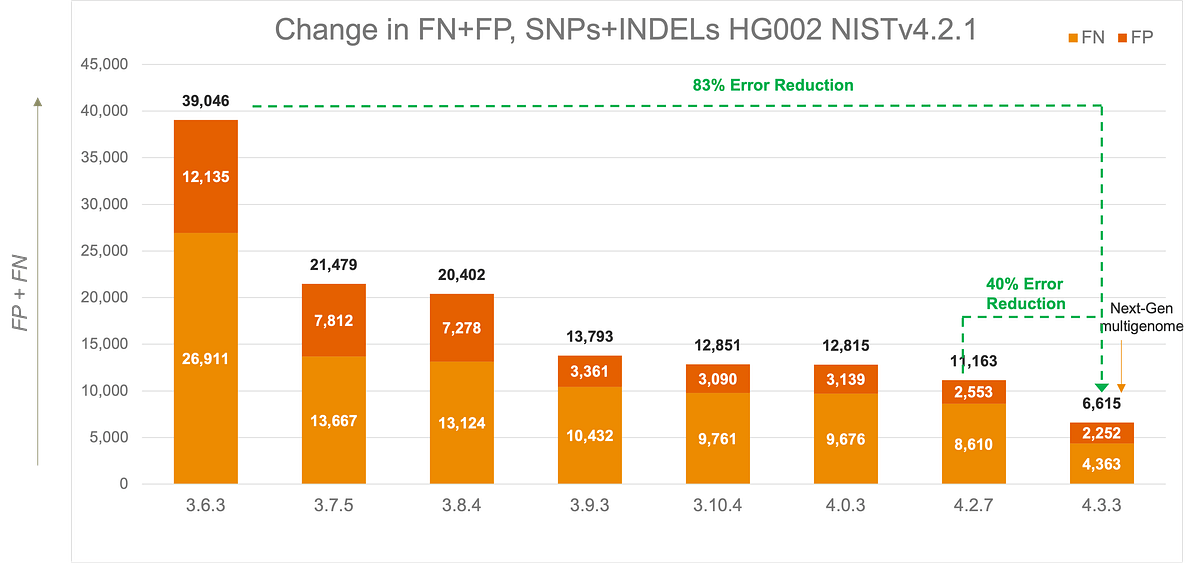
DRAGEN's multigenome (graph) mapping is now capable of utilizing a pangenome reference (also known as multigenome reference) built with few hundreds of samples. DRAGEN v4.3 comes with a set of prebuilt pangenome references from 128 samples or assemblies covering 26 ancestries. Since 2020, the DRAGEN team has invested in multigenome mapper, enabling accurate mapping in highly polymorphic regions of the genome and with this latest update, enables us to provide our most accurate secondary analysis solution to date.
Build your own custom multigenome reference
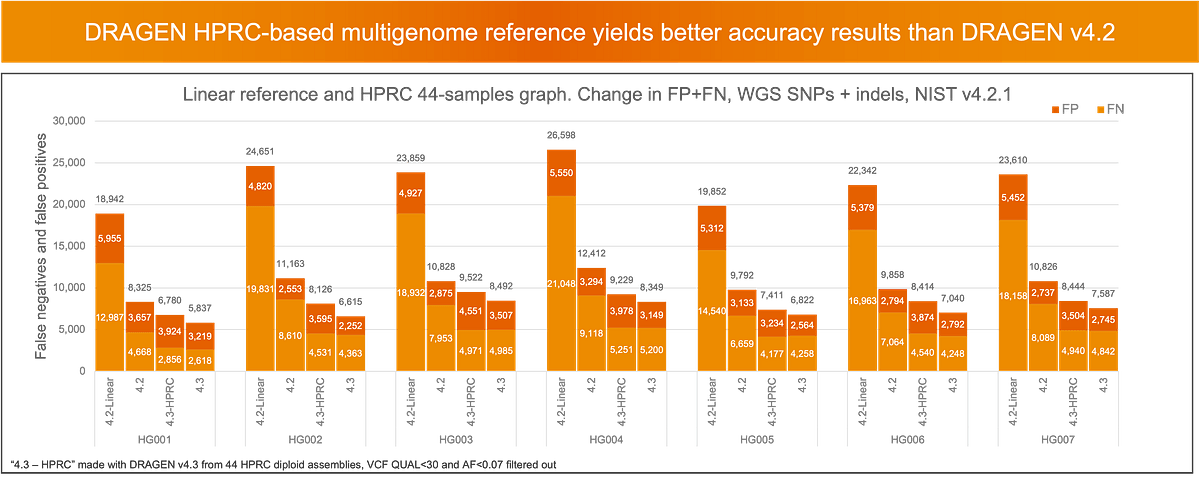
To better represent specific population and reduce ancestry bias, DRAGEN v4.3 allows users to build their own pangenome references for the DRAGEN multigenome mapper. For instance, with DRAGEN v4.3, a pangenome reference can now be built from the assemblies produced by Human Pangenome Reference Consortium (HPRC). A pangenome reference built with all 44 assemblies yields great accuracy as compared to previous versions of DRAGEN, although the default prebuilt mutigenome (pangenome) reference in DRAGEN v4.3 (based on 128 samples) still performs the best.
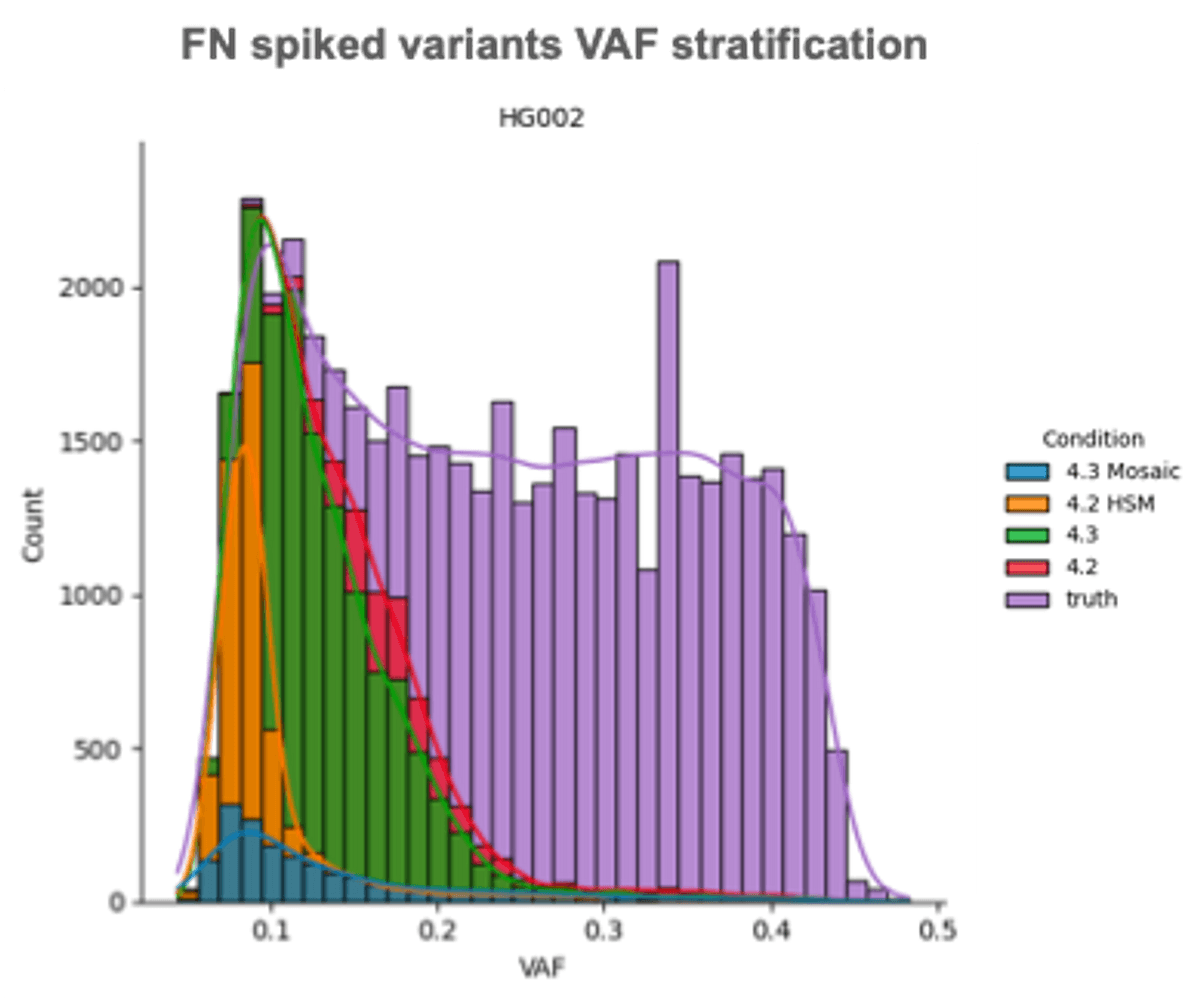
Integrated mosaic variant calling for low allele frequency variants
DRAGEN v4.3 introduces a new and improved mosaic caller. The small variant caller by default can call variants with allele frequencies greater than or equal to 20%. The mosaic mode can be optionally set to a lower threshold to detect mosaic variants at low allele frequency (less than 20%) and can be set as low as needed (e.g. <1%) to accommodate high-depth samples.
New specialized callers to scale to the long tail of problematic genes
In addition to the advanced targeted callers that are available in DRAGEN, DRAGEN v4.3 brings a new class of callers that enable de novo variant detection in regions with segmental duplications. These include:
- Multi-Region Joint Detection (MRJD) with novel variant discovery capability. These callers enable comprehensive genotyping of difficult genes with segmental duplications such as PMS2 for hereditary cancer, SMN1, SMN2, STRC, NEB for carrier screening research and TTN and IKBKG for newborn screening research.
- A new CNV (Copy Number Variants) SegDup Caller improves CNV detection in segmental duplication region across 43 clinically relevant genes used for translational research.
RNA enhancements for oncology research
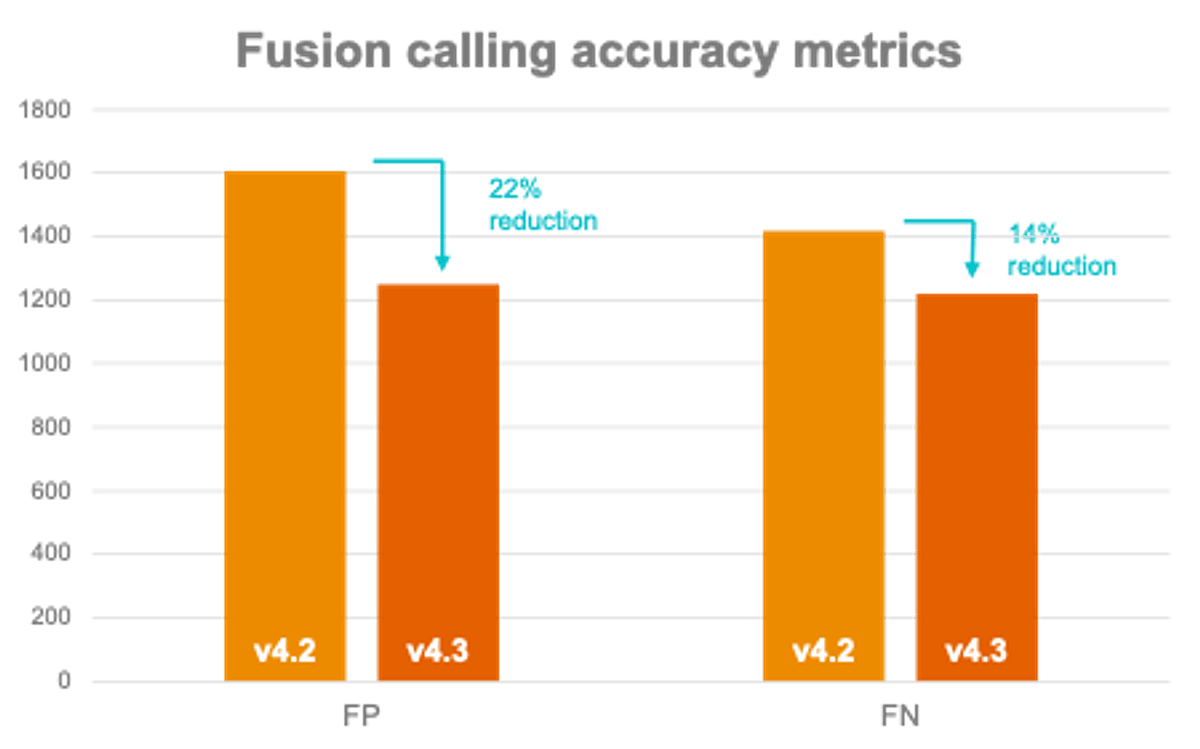
- Machine learning (ML) improvements in DRAGEN v4.3 enable more accurate gene fusion calling enhancing cancer research applications.
- DRAGEN v4.3 also comes with a new splice variant caller as an optional feature that can be enabled. This caller targets a few well-known splice variant biomarker events such as EGFR VIII, MET exon14, AR-V.
AI powered annotations
- DRAGEN v4.3 comes with Illumina Connected Annotations, which provides translational research-grade annotation of genomic variants (SNVs, MNVs, insertions, deletions, indels, STRs, gene fusions, and SVs (including CNVs). Illumina Connected annotations is the latest version of the annotation engine, incorporating PrimateAI-3D and SpliceAI as key datasets along with 20 other data sources like COSMIC, ClinVar etc. Read more here.
- Leveraging PrimateAI-3D reduces variants of unknown significance and predicts pathogenicity of all protein coding variants and SpliceAI, provides a state-of-the-art splicing prediction algorithm.
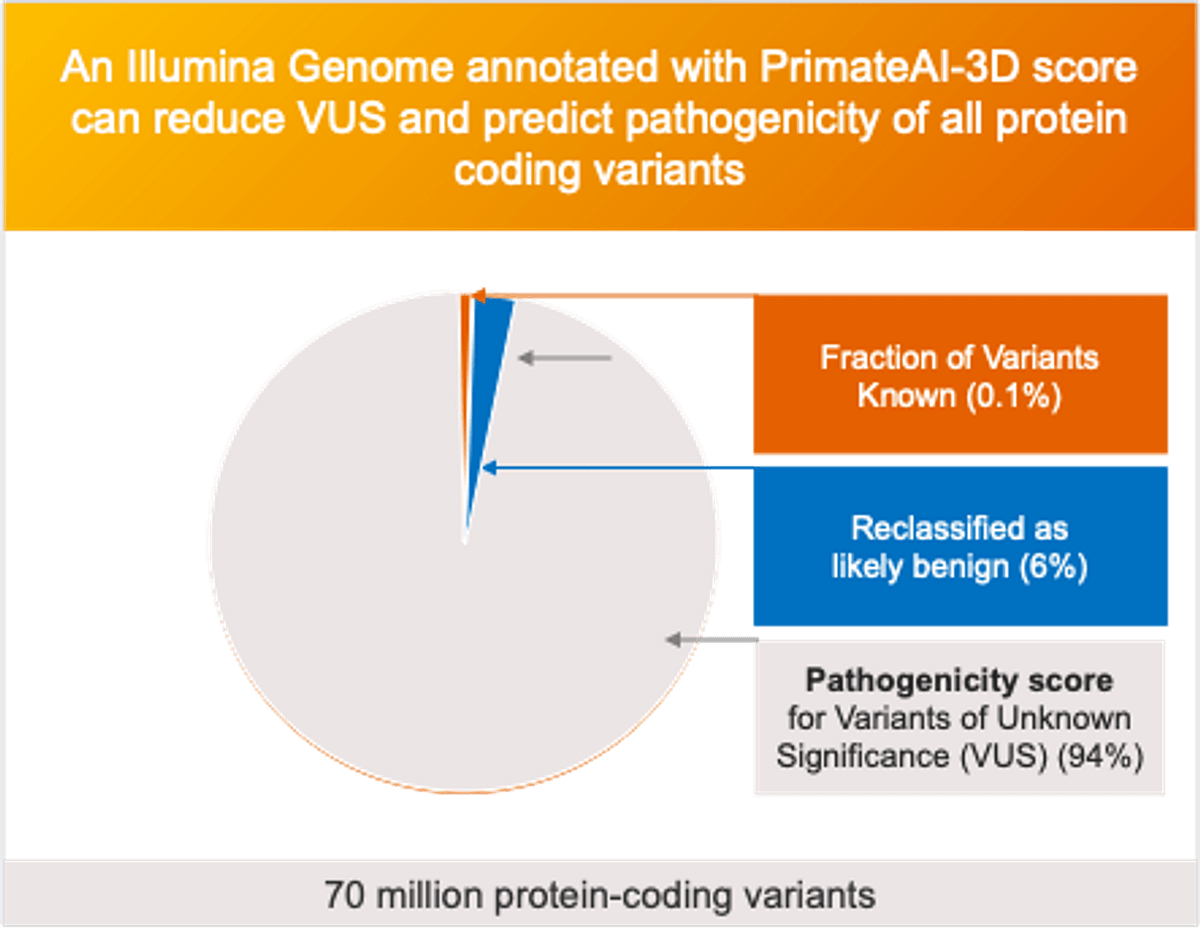
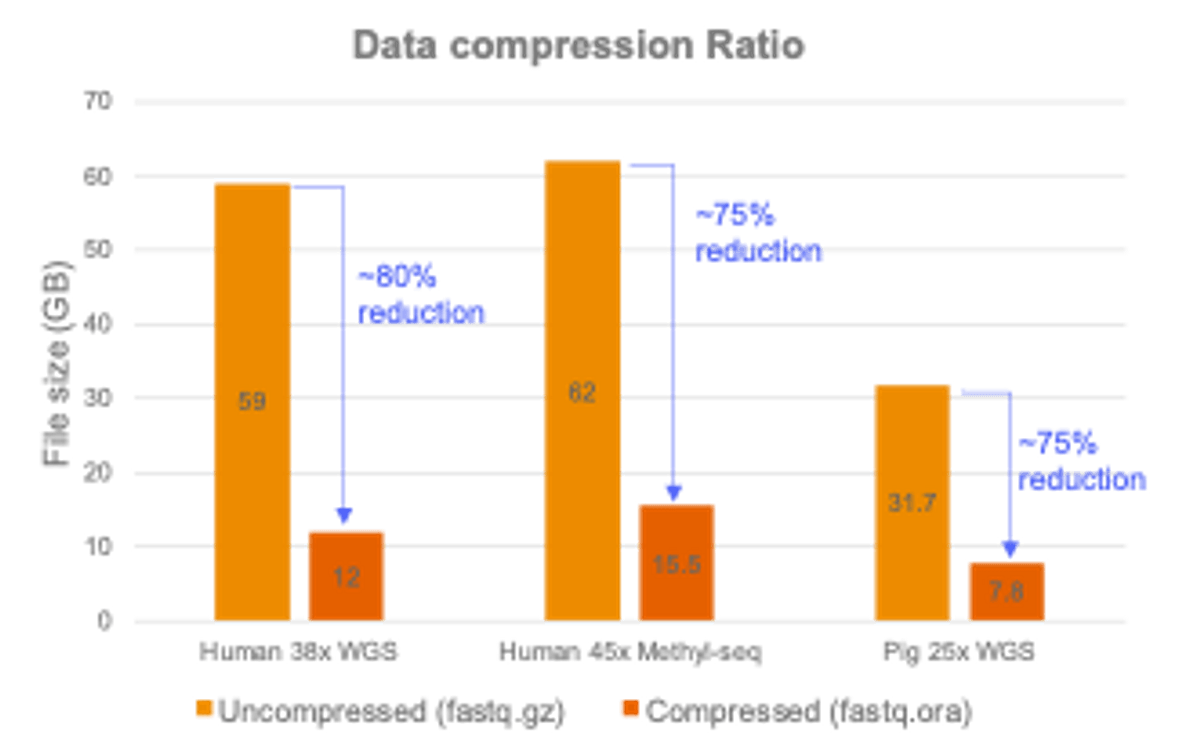
Expanded lossless Original Read Archive (ORA) compression capability, which can now support human methylation data and non-human data with a high compression ratio. Supports twelve different species including mouse, rat, zebrafish and more.
Sign up for the webinar to learn more about DRAGEN v4.3
Learn more about DRAGEN secondary analysis here
For Research Use Only
CAP# M-GL-02901