- Home
- News & Updates
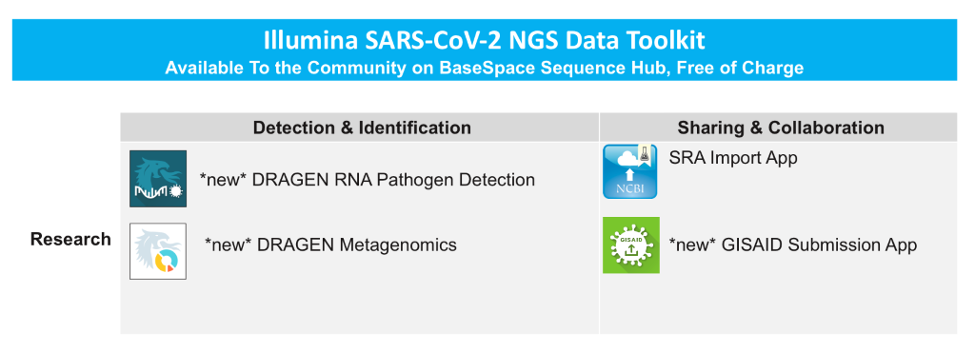
- Illumina CoV-2 NGS Data Toolkit
-
BaseSpace™ Sequence Hub
-
News
- 04/14/2020
Illumina CoV-2 NGS Data Toolkit
Illumina is committed to helping our customers across the globe address the challenges of the 2019-nCoV outbreak with a combination of world-class instruments, reagents, validated workflows, and robust bioinformatics.
To date, Illumina has released a comprehensive workflow for detecting coronavirus on our benchtop systems and announced a partnership with IDbyDNA to enable labs to develop tests that will concurrently provide comprehensive pathogen identification and information regarding antimicrobial resistance markers.
Today, we’re taking another step forward with the announcement of the Illumina SARS-CoV-2 NGS Data Toolkit, now available to the community on BaseSpace Sequence Hub, free-of-charge for all registered users. The new toolkit is comprised of several detection and identification tools built on the Illumina DRAGEN Bio-IT Platform, and data submission apps which enable researchers to seamlessly submit their findings to public databases.
In this post, I will detail the new DRAGEN COVID-19 tools released as part of the toolkit.
DRAGEN RNA Pathogen Detection Pipeline
As part of the toolkit, Illumina has released a RNA transcript analysis pipeline to enable streamlined detection of viral pathogens using coverage- and k-mer-based approaches. The new pipeline, DRAGEN RNA Pathogen Detection, enables the detection of SARS-CoV-2 in any DRAGEN RNA-seq Pipeline run, regardless of application.
To create the new pipeline, we began by leveraging the existing functionality of the DRAGEN RNA-seq (splicing-aware) aligner, as well as RNA-specific analysis components for gene expression quantification and gene fusion detection. DRAGEN uses hardware accelerated algorithms to accurately map and align RNA-Seq reads very fast – it can align 100 million paired-end RNA-Seq–based reads in about three minutes.
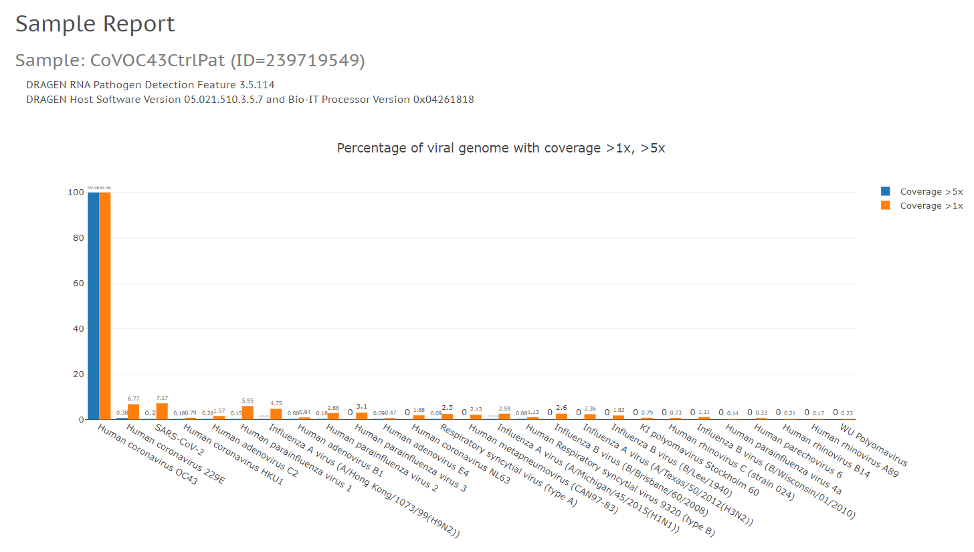
We’ve modified the DRAGEN RNA pipeline to detect SARS-CoV-2 in samples in several ways. First, we constructed a custom reference that combines human hg38 with 168 viral sequences from the Seattle Flu Study and other SARS-CoV-2 sequences. Once alignment is complete, additional post-processing is done on these results to remove duplicates and low-quality reads that are ambiguously aligned to either human or viral reference sequences. Coverage plots are then created to detect SARS-CoV-2 and other viral strains.
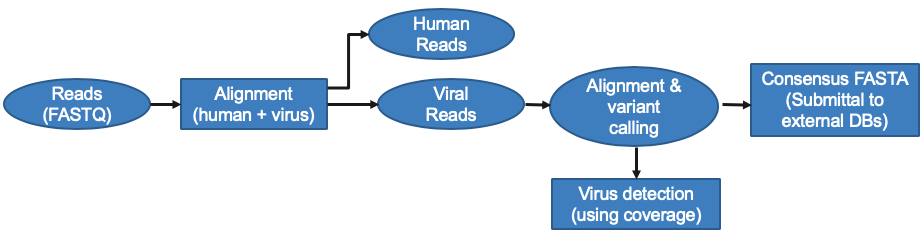
We’ve also added a custom reference based on the Illumina Respiratory Virus Panel, enabling enhanced analysis of that new panel. Custom references based on other panels or databases can also be added by customers through the app input form. Additional features are included to support variant calling and creation of consensus FASTA files for upload to public databases, such as GISAID.
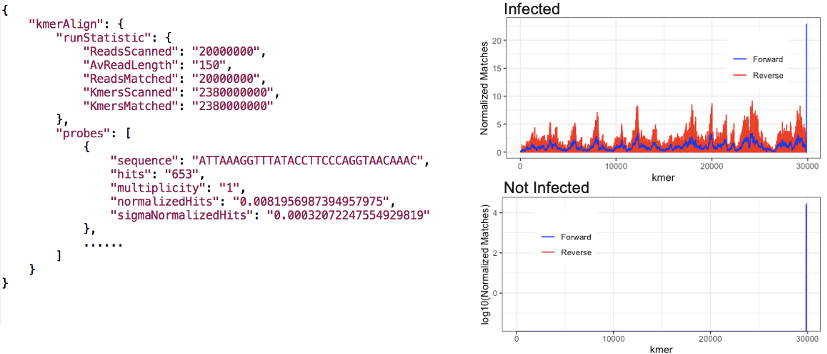
At the prompting of Nobel laureate Prof. Andrew Fire, we are adding a capability to detect SARS-CoV-2 in any DRAGEN RNA-seq pipeline run, regardless of application, and alert the operator with reporting guidance. This method scans all reads for exact matches to a set of k-mers (subsequences of length k contained within a biological sequence) specific to SARS-CoV-2 in a manner that has very little speed cost, without affecting the output of the underlying pipeline. This powerful k-mer matching engine has many possible applications. The results of this background detection process are a count of the detected k-mers and a plot of the counts.
The DRAGEN RNA Pathogen Detection Pipeline is now available on BaseSpace Sequence Hub at no-cost for the next 6 months. The team is working to deliver the new pipeline to the DRAGEN Server and DRAGEN API. Please note that an active DRAGEN annual license is required to run these tools on the DRAGEN Server.
Finally, in addition to modifications of the existing DRAGEN RNA pipeline, we are also excited to announce the release of DRAGEN Metagenomics, a k-mer based classification workflow that is able to detect and quantify SARS-CoV-2 sequences at high sensitivity and specificity while simultaneously providing readouts for other common viral and microbial pathogens.
DRAGEN Metagenomics Pipeline
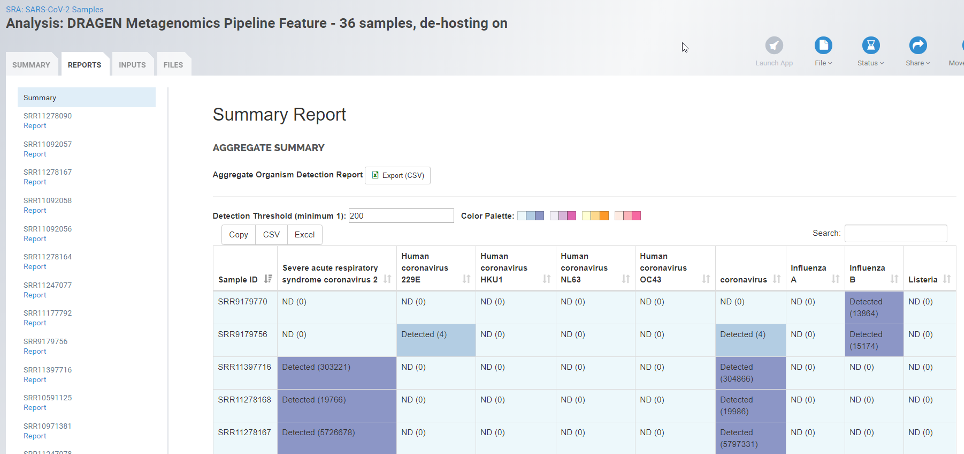
The new DRAGEN Metagenomics Pipeline takes advantage of DRAGEN Aligner to remove host reads, which is an important step in many metagenomics applications. Sequences contributed by the microbes of interest are vastly outnumbered by sequences from the host organism. In addition to increasing processing time, the presence of such sequences can confound downstream applications such as classification and genome assembly. The unparalleled speed of DRAGEN enables accurate removal of host sequences with negligible run-time penalty.
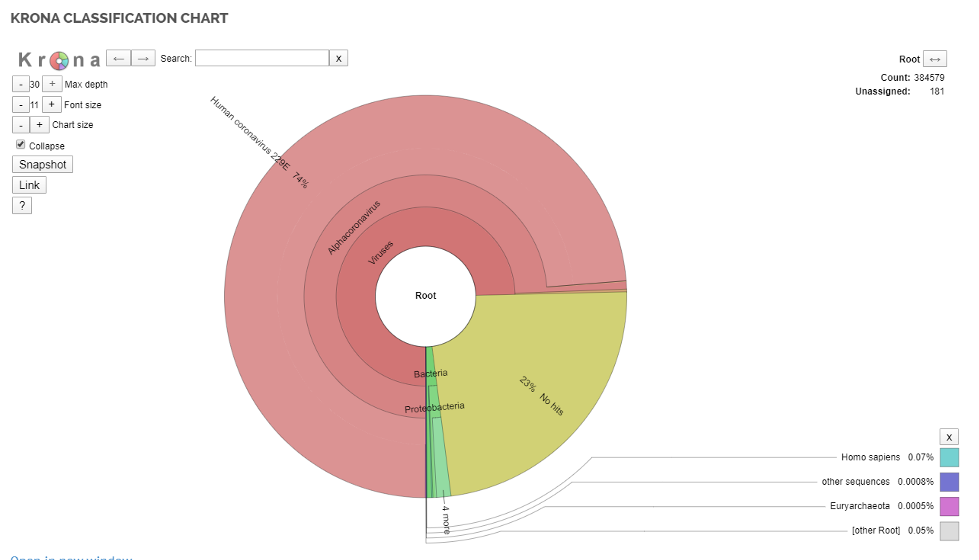
Once data are “de-hosted,” the pipeline leverages Kraken2 , a best-in-class metagenomics classification algorithm, to count unique, diagnostic k-mers and estimate the relative abundance of the organisms present in its database. Kraken2 is currently a preferred tool for researchers investigating metagenomics, microbiomes and viral genomics around the world, and integrating it within the larger DRAGEN Bio-IT Platform enables more accurate and faster analysis than was previously available.
Data Sharing Apps
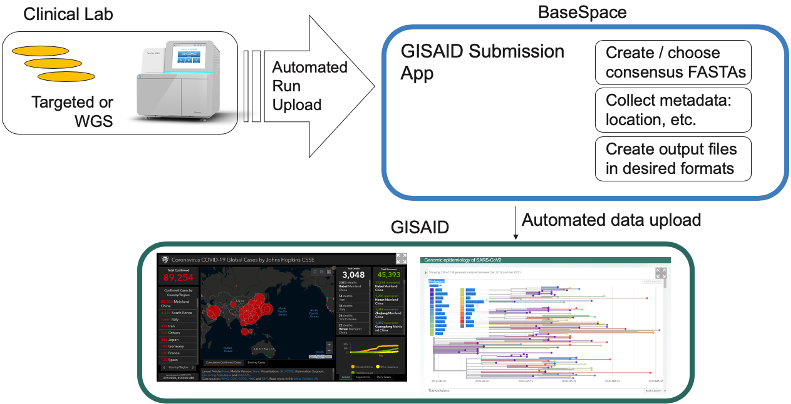
To enable simple data sharing, Illumina has also released two data sharing apps, enabling push-button submissions to GISAID (Global Initiative on Sharing All Influenza Data) and the NCBI SRA (Short Read Archive).
Once data are analyzed, researchers can seamlessly contribute sequences to central resources to enable outbreak surveillance and other epidemiological analyses. Illumina BaseSpace Sequence Hub is also releasing the Submission App and updated applications for data submission to and importing from the NCBI SRA.
Getting Access to the Toolkit
Researchers can start using the Illumina SARS-CoV-2 NGS Data Toolkit today on BaseSpace Sequence Hub, free of charge until Oct. 31, 2020. Researchers can stream data directly from their instruments into BaseSpace’s secure cloud-environment for push-button usage of the entire toolkit.
In addition to BaseSpace Sequence Hub, we will provide Illumina DRAGEN Server customers a special build of DRAGEN version 3.5 that has the RNA and k-mer pipeline enhancements on our DRAGEN support page in May 2020. Finally, we are pleased to announce that these pipelines will also be made available within a preview of a new Platform-as-a-Service offering, DRAGEN API, which will be made available in May. DRAGEN API is built on top of Amazon Web Services and allows users to call a simple API endpoint to stream, process, and deliver COVID-19 sample data to and from their own AWS S3 buckets. If you are interested in learning more about DRAGEN API, please fill out the form on the DRAGEN SARS-CoV-2 NGS Data Toolkit web page.
Special thanks to Eric Allen, Jay Patel, and Shyamal Mehtalia who contributed greatly to this post.