- Home
- News & Updates
- Introducing DRAGEN™ Array 1.0 for Infinium™ Array-Based Pharmacogenomics Analysis
-
DRAGEN™ Array
-
Product updates
-
News
- 12/13/2023
Introducing DRAGEN™ Array 1.0 for Infinium™ Array-Based Pharmacogenomics Analysis

Introduction
DRAGEN Array is a new bioinformatics software suite from Illumina for accurate, comprehensive, and efficient analysis of Infinium microarray data. The software is for Research Use Only (RUO).
The first release, DRAGEN Array 1.0, provides the capability for standard genotyping analysis for Infinium arrays and a pharmacogenomics (PGx) analysis pipeline for the Infinium Global Diversity Array with enhanced PGx (GDA-ePGx) arrays (Figure 1).
DRAGEN Array provides a local command line interface (CLI) and a cloud-based web user interface. DRAGEN Array Local supports both Windows and Linux operating systems. The software runs on CPUs and does not require a specialized server or FPGA hardware. DRAGEN Array Local allows users to have granular control and flexibility to support large scale microarray genomics studies, while DRAGEN Array Cloud provides predefined pipelines with a user-friendly graphical interface on BaseSpace Sequence Hub.
Pharmacogenomics
Pharmacogenomics studies the impact of individuals’ genetic variation on their responses to medicine. PGx solutions allow scientists and translational researchers to harness and translate genetic data into insights of drug efficacy, side effects, dosing, and selection. PGx focuses on the core variants that alter the functions of PGx genes. The star nomenclature provided by the Pharmacogene Variation Consortium (PharmVar) is often used for PGx gene haplotypes or “star alleles”, e.g., *1, *2, representing verified combinations of the core variants.
There are multiple challenges with the task of PGx star allele calling, including:
- High homology between the key PGx genes such as CYP2D6 and their pseudogenes,
- Platform and sample specific genotyping errors,
- Complex Structural Variations (SV) and Copy Number Variations (CNV), and
- Phasing of core variants and associations of CNV with star alleles.
Illumina provides analysis solutions to address these challenges through both array and sequencing platforms with DRAGEN Array PGx analysis and DRAGEN PGx analysis, respectively.
Below we dive into the DRAGEN Array PGx analysis.
DRAGEN Array PGx Analysis Pipeline
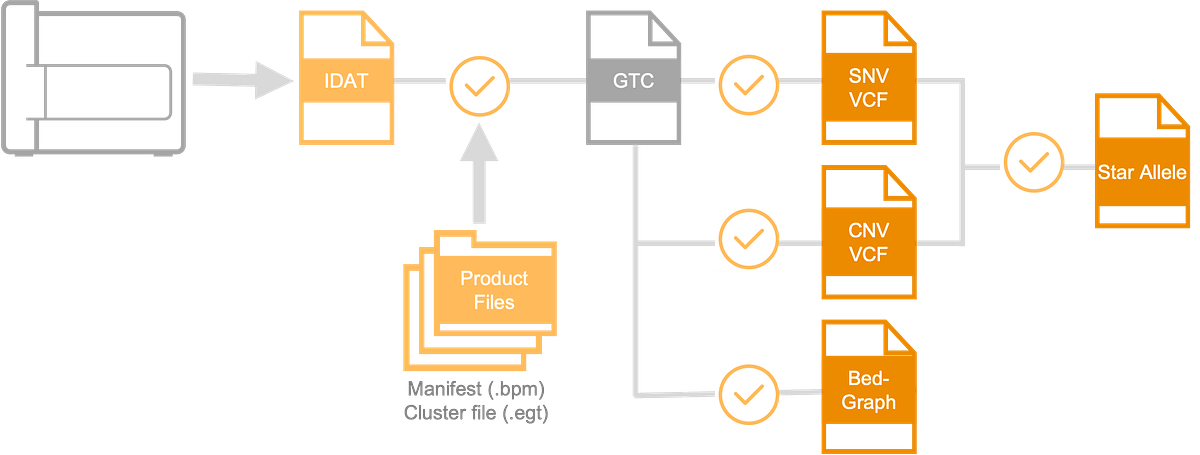
DRAGEN Array enables genotyping, PGx CNV calling, and star allele calling in a single analysis platform, with multiple output options. The DRAGEN Array PGx pipeline takes in IDAT files and produces star allele annotations in JSON file format (Figure 2).
PGx CNV Calling
DRAGEN Array is optimized to give accurate PGx CNV results for Infinium microarrays with enhanced PGx content. With a few simple commands, the software processes the IDATs raw data and determines star allele calls for hard to discern PGx genes including CYP2D6.
Star Allele Calling
DRAGEN Array provides robust star allele calls that are tolerant to input data noise and provides QC scoring and supporting variant information to enable downstream evaluation. It also provides metabolizer status annotations for star allele diplotype calls.
Genotyping Analysis
Using DRAGEN Array, small variant genotyping can be efficiently performed on any Infinium microarray, including GDA-ePGx, saving time for large experiments.
Standardized Output
DRAGEN Array produces Variant Call Format (VCF) outputs for both genotyping and PGx CNV calling, which can then be incorporated into further multi-omic analyses. DRAGEN Array star allele calls and phenotype annotations are captured in CSV and JSON format outputs for human-readability and easy integration into optional downstream clinical interpretation pipelines provided by third parties.
DRAGEN Array PGx Analysis Capability
Comprehensive PGx Gene Coverage
DRAGEN Array PGx analysis integrates curated and peer-reviewed knowledge from public databases including the Clinical Pharmacogenetics Implementation Consortium (CPIC), Pharmacogene Variation (PharmVar) and Pharmacogenomic Knowledge Base (PharmGKB) to provide comprehensive coverage of key PGx genes. Table 1 provides details on specific genes that are covered by the DRAGEN Array PGx caller.
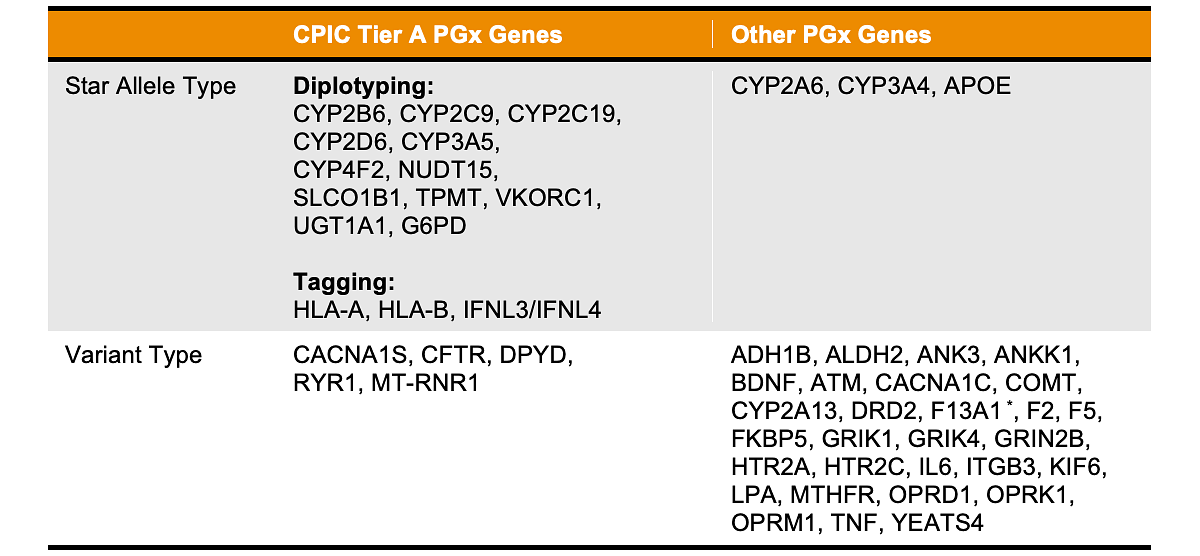
PGx genes covered by DRAGEN Array 1.0 fall into two categories:
- Variant Type: These genes are characterized by individual variants genotyped directly from the array data by the DRAGEN Array PGx pipeline.
- Star Allele Type: For 4 genes (HLA-A, HLA-B, IFNL3/IFNL4), tagging variants are used to infer the star alleles or phenotypes of the genes. All other genes in the category are subjected to direct star allele diplotyping, using the variants detected on the array to determine the star allele combinations in each sample. A star allele combination could involve hybrid gene alleles and copy number changes of star alleles including gene deletions, duplications and multiplications.
Native Star Allele Calling
DRAGEN Array 1.0 presents a new and native PGx star allele caller with multiple unique features compared to other PGx solutions. The software:
- Is tolerant to missing data from array genotyping and reports missing variants and affected star alleles (see Figure 3 for an example JSON output for CYP2D6).
- Provides variant level supporting information such as genotype calls and GS scores.
- Provides star allele-specific copy numbers.
- Provides quality scores associated with determined star allele diplotypes, which can be used to assess the star allele calls in downstream analysis.
- Provides activity score in addition to metabolizer status annotations for three key PGx genes (CYP2C9, CYP2D6, and DPYD).
- Provides ranked alternative star allele calls for a sample to enable downstream manual evaluation or experimental assessment when needed.

DRAGEN Array PGx caller replaces PGx analysis + interpretation v1.1.1, which was integrated with Illumina’s cloud platform in 2021. DRAGEN Array provides an improved PGx call rate, with less than 2% no-call rate compared to 8%-20% for the previous solution.
PGx CNV Calling and Genotyping Performance
DRAGEN Array 1.0 provides accurate genotyping and CNV calls. On a dataset of 384 Coriell samples run using Global Diversity Array with enhanced PGx, greater than 99% genotyping accuracy was achieved on key PGx content and for all variants covered by GDA-ePGx (Table 2) when compared against NGS references.

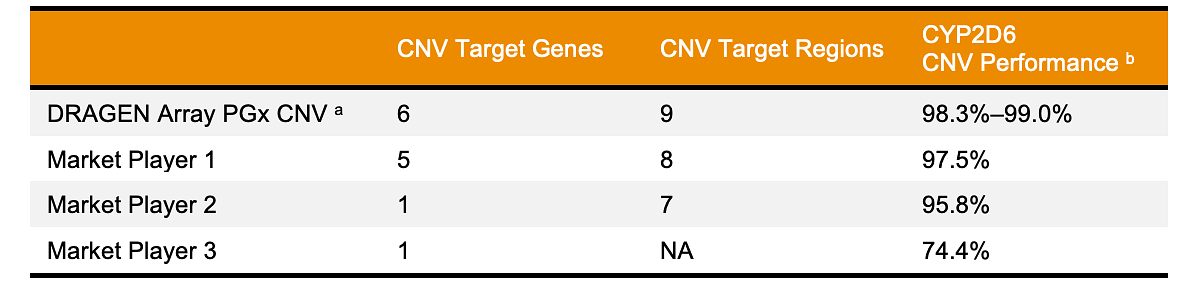
DRAGEN Array demonstrates an overall accuracy of 98.3%–99.0% for CYP2D6 on a set of 288 diverse samples against q-PCR reference copy number calls (Table 3). Greater than 95% accuracies were achieved across 9 CNV target regions on multiple datasets.
PGx Star Allele Calling Performance
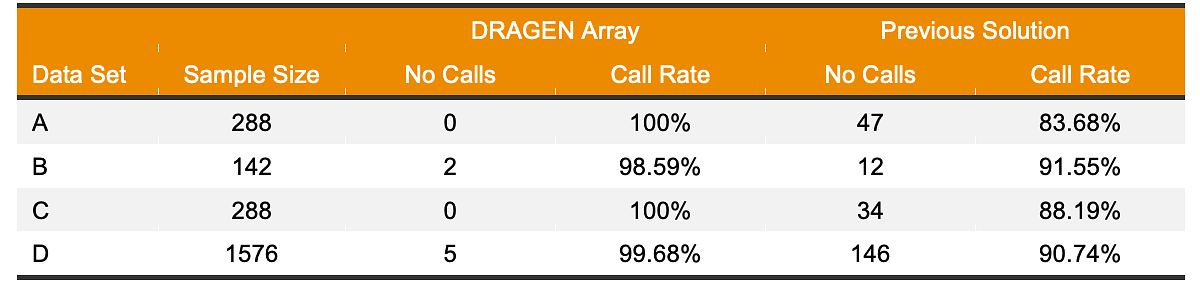
Besides better gene coverage and accuracy, DRAGEN Array provides significant improvements over the previous star allele calling solution in terms of star allele call rate. DRAGEN Array PGx caller calls more star alleles with fewer no calls (indeterminants). Table 4 shows the call rate performance of DRAGEN Array PGx on the gene CYP2D6, which is one of most important and challenging PGx genes to test. DRAGEN Array PGx caller demonstrated > 98% call rate, while the previous solution shows call rates of 80%–92%.
Conclusion
DRAGEN Array is a software solution for analyzing Illumina array data. DRAGEN Array 1.0 delivers unique PGx analysis capabilities to enable comprehensive and accurate PGx CNV calling and star allele annotation. It provides standardized and information-rich output files for integration with downstream PGx evaluation and translational research pipelines.
Learn More
DRAGEN Array product page on illumina.com
Contact Illumina Customer Care
For Research Use Only. Not for use in diagnostic procedures.
M-GL-02484