- Home
- News & Updates
- MiXCR Immune Repertoire Analyzer version 2.1.11 from MiLaboratories is now available at Illumina BaseSpace™
-
BaseSpace™ Sequence Hub
-
News
- 11/21/2019
MiXCR Immune Repertoire Analyzer version 2.1.11 from MiLaboratories is now available at Illumina BaseSpace™
- Youting Sun, Senior Bioinformatics Scientist at Illumina
- Dmitriy Chudakov, CSO at MiLaboratories https://milaboratory.com
The upgraded version of the MiLaboratories LLC flagship software product, MiXCR, is now officially available as an Illumina BaseSpace™ application.
MiXCR is a “gold standard” analytical package in the area of T-cell receptors (TCR) and immunoglobulin (IG) repertoire profiling. Analysts apply MiXCR for extracting immune repertoires from any type of sequencing data with any level of TCR/IG coverage, ranging from perfectly enriched libraries such as multiplex PCR or targeted 5’RACE, to the “rare event” datasets containing several target entities among hundreds of millions of reads, such as RNA-Seq, and even Exome-Seq data.
In the new version, simple selection of species (Human or Mouse), template material (RNA or DNA), library type (targeted or random), and basic information on library preparation enables appropriate analysis settings for a variety of immune repertoire experiment scenarios:
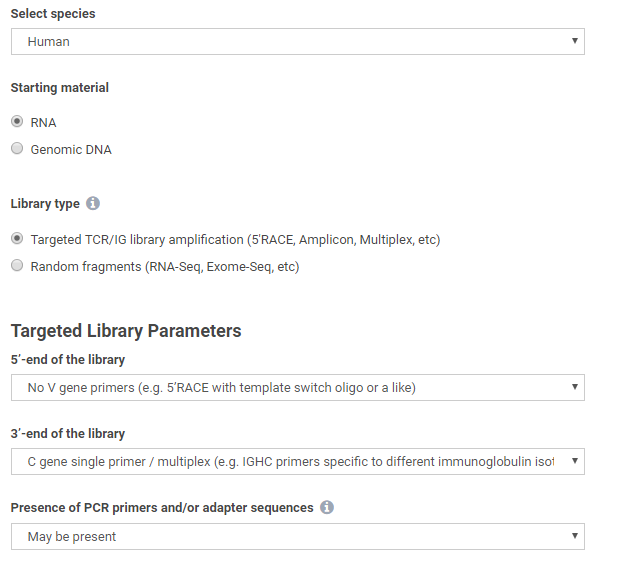
Extraction of both full VDJ length or CDR3 only repertoires is possible, for TCR or IG chains of interest, with or without out-of-frame and stop codon-containing clonotype variants:
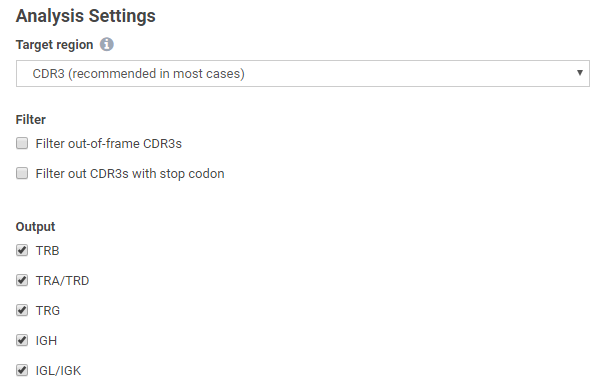
The application also provides post-analysis metrics in the form of interactive reports, including:
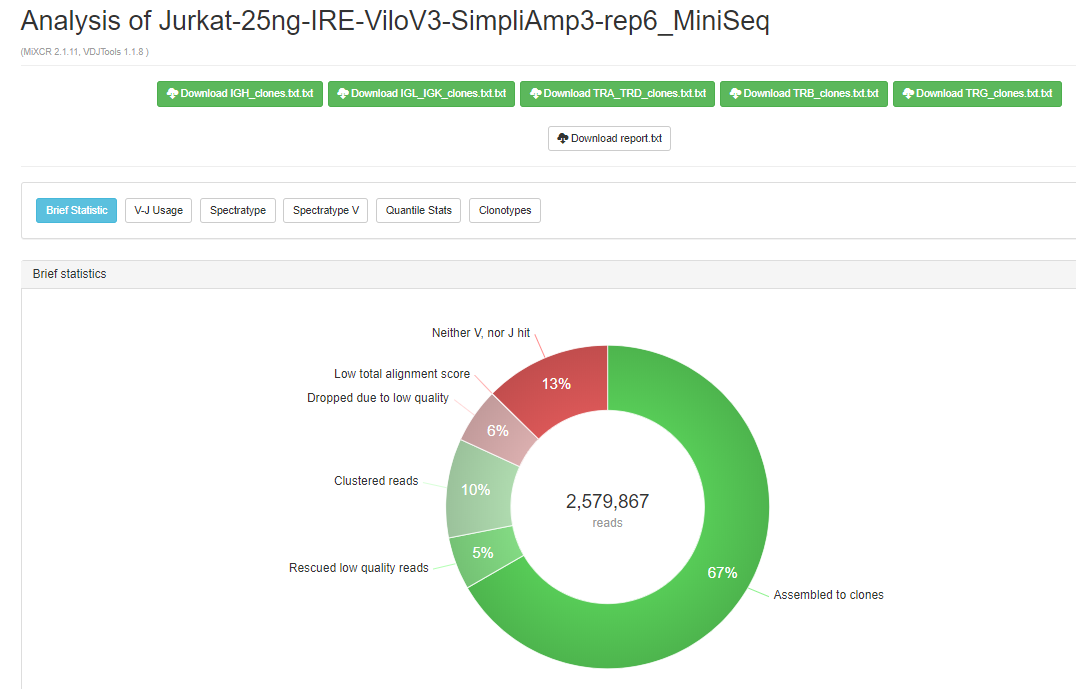
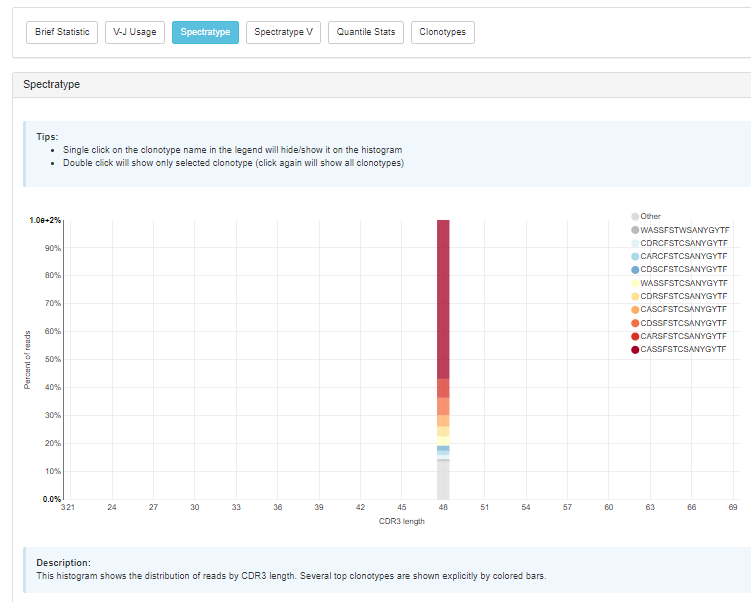
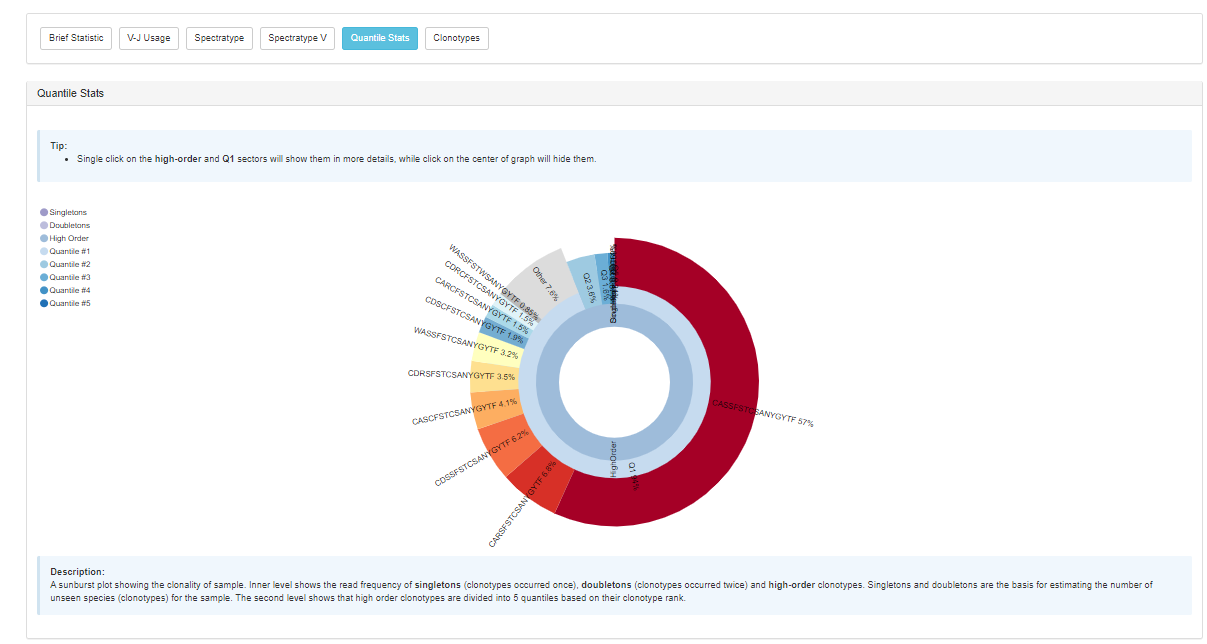
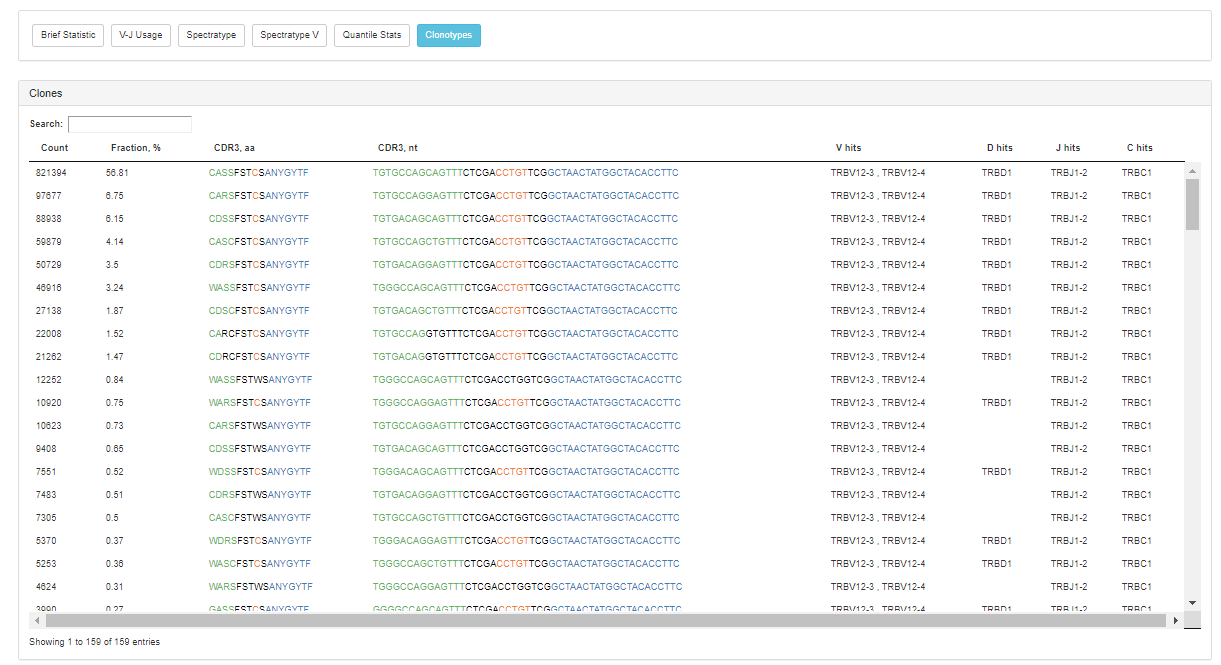
High extraction efficiency for any type of sequencing data and superb accuracy should make the new MiXCR BaseSpace version a highly useful resource for many. The ability to look at basic parameters and immediately download the resulting figures for reports and publication make it really convenient for the most efficient everyday work on immune repertoires.
Analysis of amplicon data
Recommended settings for panels (a) Immune repertoire panel and (b) TCR-beta SR panel:
- Starting material: RNA for panel (a), RNA or Genomic DNA for panel (b)
- Library type: Targeted TCR/IG library amplification (5'RACE, Amplicon, Multiplex, etc)
- 5’-end of the library: V gene single primer/multiplex
- 3’-end of the library: J gene single primer/multiplex
- Presence of PCR primers and/or adapter sequence: Absent / nearly absent / trimmed
- Target region: CDR3
- Demo datasets:
- MiniSeq: Immune Repertoire Panel (Replicates of Jurkat)
- NextSeq 550: Immune Response Panel (UHR, Brain, Lung, and HL60)
- NextSeq: TCR beta-SR Panel (Replicates of Jurkat and PBMC)
For Research Use Only. Not for use in diagnostic procedures.
QB#9017