- Home
- News & Updates
- Q2 2020 | Illumina Informatics Quarterly Updates
-
DRAGEN
-
News
- 06/15/2020
Q2 2020 | Illumina Informatics Quarterly Updates
We hope you and your family, community and colleagues are staying safe. Before we detail our product updates for the second quarter of 2020, we wanted to thank all of the researchers committed to the fight against COVID-19.
To learn more about how Illumina is supporting the COVID-19 research community, visit our website, and read below to learn more about the new COVID-19 tools available to the research community, free-of-charge, through our BaseSpace™ Suite.
Upcoming Webinars:
- Illumina DRAGEN Bio-IT Platform: What’s new with version 3.6
- Wednesday, June 24 at 8 a.m. PDT
- Registration is now live
- Our Magic DRAGEN: A Sequencing Lab Case Study on Accelerated Bioinformatics. In this webinar, Dr. Charlie Johnson, founder of the Texas A&M AgriLife Genomics and Bioinformatics Service, will share how his team is utilizing Illumina’s DRAGEN informatics platform in its high-throughput agrigenomics research program.
- Thursday, July 9 at 1 p.m. EDT
- Registration is now live
Software Tools for the COVID-19 Research Community
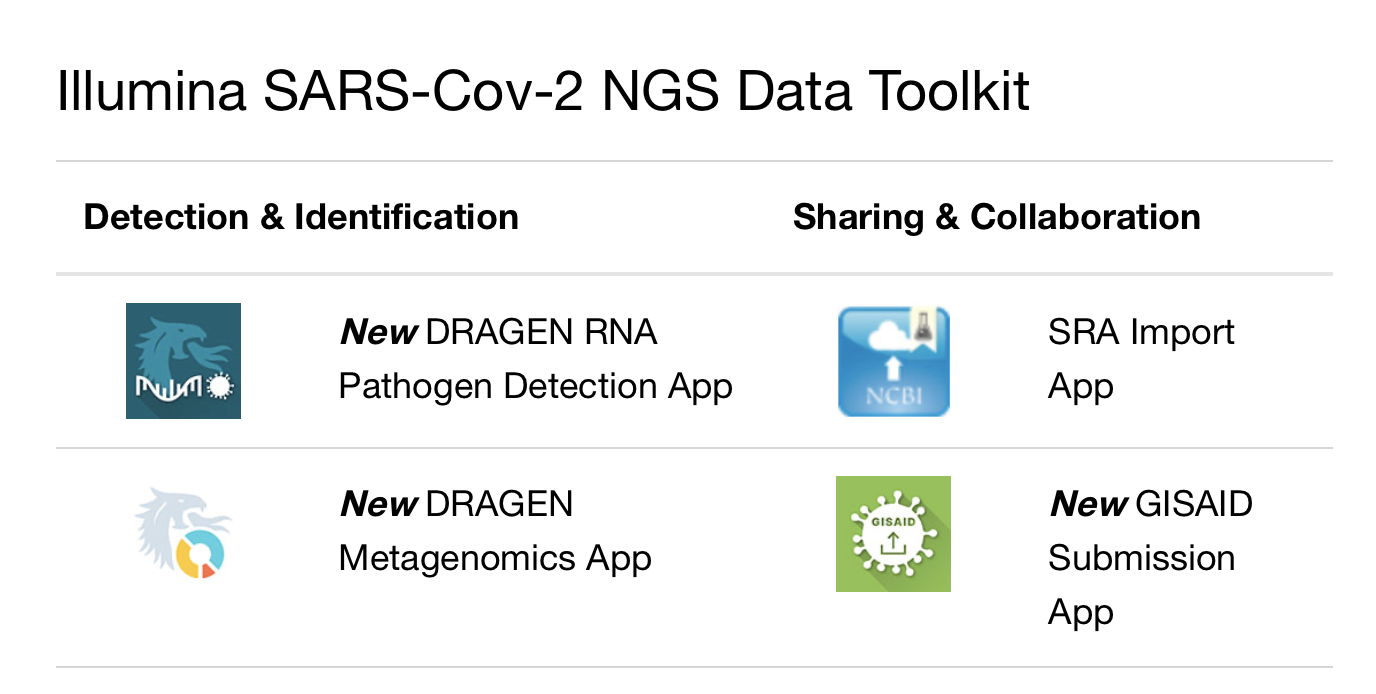
To support researchers with analysis and sharing of genomic data in relation to the severe acute respiratory syndrome coronavirus outbreak, Illumina has released the Illumina SARS-CoV-2 NGS Data Toolkit – a new suite of data analysis tools and functionality for scientists working with the virus.
These tools make it simpler for researchers to detect and identify the SARS-CoV-2 viral sequence in their samples, and contribute their findings to critical public databases. Learn more about the new DRAGEN Apps released in the Illumina SARS-CoV-2 NGS Data Toolkit. This coronavirus detection and identification software is available to the community free of charge on BaseSpace Sequence Hub.
Additionally, we've released two new BaseSpace Clarity LIMS workflows to accelerate comprehensive workflow management and sample tracking for COVID-19:
- CDC COVID-19 RT-PCR workflow based on CDC recommendations
- Respiratory Virus Panel workflow using Nextera Flex for Enrichment library prep kit
Learn more about the new Clarity LIMS COVID-19 workflows.
New Regional Instance|Canada
llumina's BaseSpace Sequence Hub is now commercially available in Canada, coupling the scalability and cost-efficiency of Illumina's cloud based compute and storage environment with in-region data hosting. Hosted on a new Amazon Web Services (AWS) regional instance in Montreal, BaseSpace Sequence Hub Canada complies with data sovereignty requirements and security regulations, including PIPEDA and PHIPA.
Learn more.
Genomic Lab Workbench
BaseSpace Clarity LIMS: v5.3 featuring Illumina Preset Protocols, NextSeq 2000 Integration and more
We are pleased to announce the launch of BaseSpace Clarity LIMS with version 5.3. Clarity LIMS is a cloud genomics LIMS that provides regulated labs with efficient sample tracking and workflow management. Key features include:
- Illumina Preset Protocols: out-of-the-box compatibility and integration with Illumnia’s library preparation portfolio and sequencers
- Support for Illumina’s NextSeq™ 2000: Seamless compatibility and Illumina’s newest sequencer
- Security Improvements
Genomic Data Workbench
Something to Celebrate: DRAGEN 2-Year Anniversary
It has been two-years since Illumina acquired Edico Genome and its DRAGEN™ technology. Since then, we have seen new developments, partnerships and advances all centered around a singular vision – a commitment to delivering value to our customers. Two-years in, we’re excited to reflect back on our growth and explore how we’re enabling researchers to ask new questions and glean more valuable data with our expanded portfolio.
New DRAGEN RNA Pathogen Detection and DRAGEN Metagenomics Pipelines
As part of the Illumina SARS-CoV-2 NGS Data Toolkit, Illumina has released a RNA transcript analysis pipeline to enable streamlined detection of viral pathogens using coverage- and k-mer-based approaches. The new pipeline, DRAGEN RNA Pathogen Detection, enables the detection of SARS-CoV-2 in any DRAGEN RNA-seq Pipeline run, regardless of application.
The new DRAGEN Metagenomics Pipeline takes advantage of DRAGEN Aligner to remove host reads, which is an important step in many metagenomics applications. Sequences contributed by the microbes of interest are vastly outnumbered by sequences from the host organism. In addition to increasing processing time, the presence of such sequences can confound downstream applications such as classification and genome assembly. The unparalleled speed of DRAGEN enables accurate removal of host sequences with negligible run-time penalty.
Learn more about the new pipelines.
BaseSpace Variant Interpreter Updates
Interpret the DRAGEN: We released support for the DRAGEN somatic pipeline in BaseSpace Variant Interpreter. This means oncology researchers will now be able to harness the speed, accuracy, and scalability of DRAGEN alongside the paralleled benefits of BSVI, enabling rapid secondary and tertiary analysis. DRAGEN results may be imported into BSVI directly from BaseSpace Sequence hub, or uploaded to BSVI using the VI upload capabilities, for users with an on-premise DRAGEN server
Other BSVI Improvements: BSVI will also receive a number of other improvements for Q2, including updates to the annotation engine and data sources. Read the recent release notes.
For research use only. QB #9989