- Home
- News & Updates
- VCAT v4.0 - Support for DRAGEN and GA4GH best practices for small variant benchmarking
-
BaseSpace™ Sequence Hub
-
DRAGEN
-
News
- 11/26/2019
VCAT v4.0 - Support for DRAGEN and GA4GH best practices for small variant benchmarking

Author: Eric Allen, Associate Director of Bioinformatics at Illumina
The Variant Calling Assessment Tool (VCAT) v4.0 BaseSpace™ App has an updated hap.py and several new gold references and panel BED files.
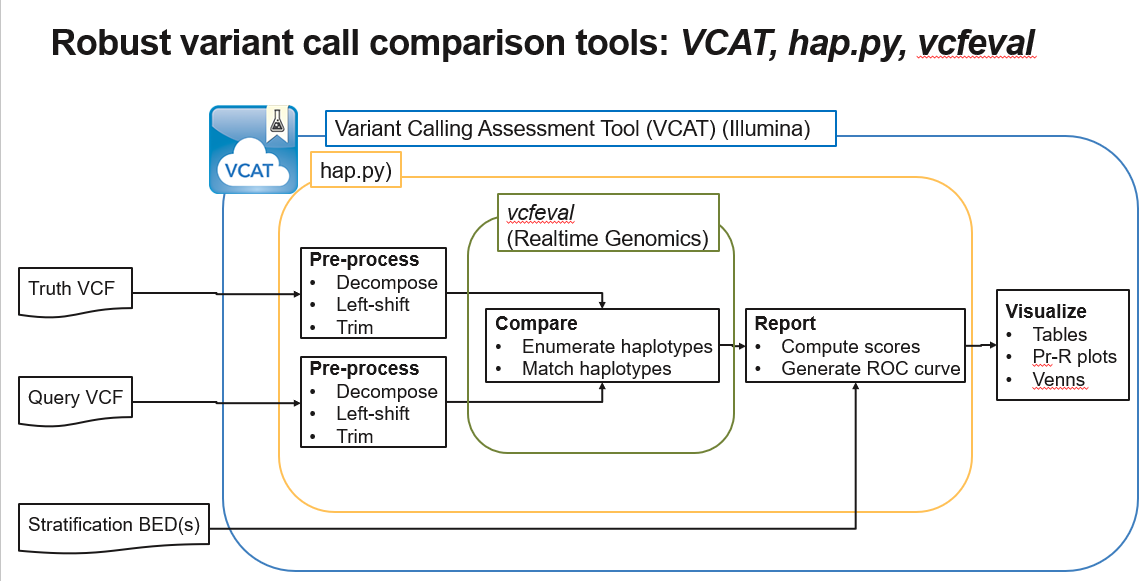
This update includes a recent version of hap.py which (in combination with vcfeval) has been selected by the GA4GH as the recommended tool for small variant call benchmarking. For more details, see the publication at: https://www.ncbi.nlm.nih.gov/pubmed/30858580
Updated and improved tools
- hap.py 0.3.12, which uses the
vcfevalcomparison engine from rtg-tools. - Improved handling of DRAGEN™ and GATK VCF and gVCF files
- Added default filtering of variants genotyped as <NON_REF>
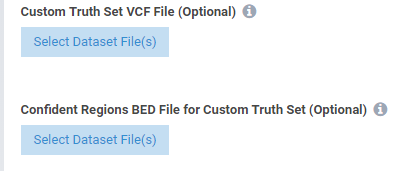
- Added ability to import custom truth sets
Gold Reference Additions and Updates
- Updated Platinum Genomes reference data to version 2017-1.0
- Added all the NIST GiaB gold references to v3.3.2:
- Caucasian female: NA12878 (mother)
- Ashkenazi Jewish trio: NA24143 (mother), NA24149 (father), NA24385 (son)
- Han Chinese trio: NA24695 (mother), NA24694 (father), NA24631 (son)
More built-in panel BED files
- Illumina, IDT, and Twist exome panels
- Several AmpliSeq panels
The diagram below shows how VCAT, hap.py, and vcfeval work together.
See the screenshots below displaying some of the new features.

We hope you find VCAT 4.0 useful. Questions can be directed to basespacelabs@illumina.com.
For Research Use Only. Not for use in diagnostic procedures.
QB#9047