- Home
- News & Updates
- 10x Genomics Chromium Single Cell Multiome ATAC + Gene Expression data now available on Illumina® BaseSpace™ Sequence Hub
-
BaseSpace™ Sequence Hub
-
News
- 06/21/2021
10x Genomics Chromium Single Cell Multiome ATAC + Gene Expression data now available on Illumina® BaseSpace™ Sequence Hub
Today, we’re sharing the third in a series of posts that will shed more light on new run data publicly available on BaseSpace™ Sequence Hub (BSSH).
Single-cell analysis can help tease apart heterogeneity in complex cell populations, distinguishing cell types and revealing dynamic cell states. 10x Genomics is a partner that develops multiple single cell and spatial biology solutions. The Chromium Single Cell Multiome ATAC + Gene Expression product (“10x Genomics Multiome product”) prepares sequencing-ready libraries to simultaneously profile gene expression and chromatin accessibility from single cells.
Today, we are excited to announce that we have published sequencing data for this product on BaseSpace™ Sequence Hub, including one set of libraries sequenced on the NextSeq™ 2000 P2 flow cell, and another set of libraries sequenced on a NovaSeq™ 6000 SP v1.5 flow cell.
10x Genomics Multiome assay workflow
The 10x Genomics Multiome assay distinguishes itself from other experimental designs that combine multiple “omes” by using the single-cell barcodes to directly link chromatin accessibility data to gene expression data. This is achieved by generating both ATAC and 3' gene expression libraries from the same starting material.
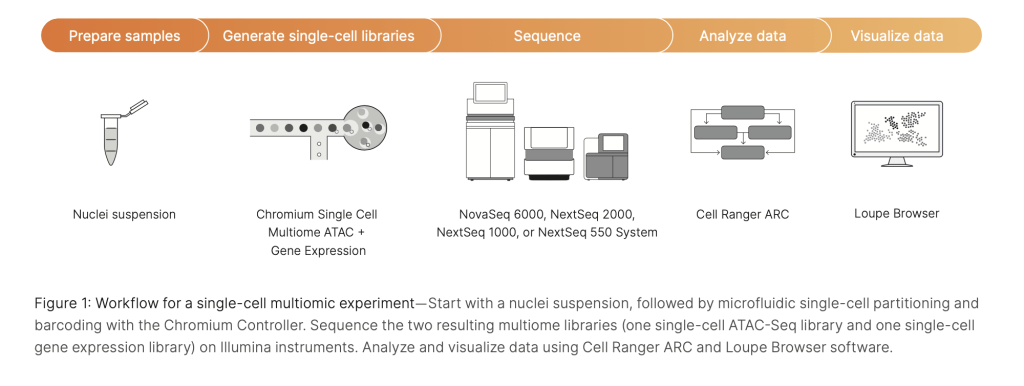
The overall workflow for preparing 10x Genomics Multiome libraries starts with a nuclei suspension, followed by microfluidic single-cell partitioning and barcoding with the Chromium Controller. Sequencing for the two resulting Multiome libraries (one single-cell ATAC-Seq library and one single-cell gene expression library) then happens on Illumina® instruments, followed by analysis and data visualization using 10x Genomics Cell Ranger ARC and Loupe Browser software.
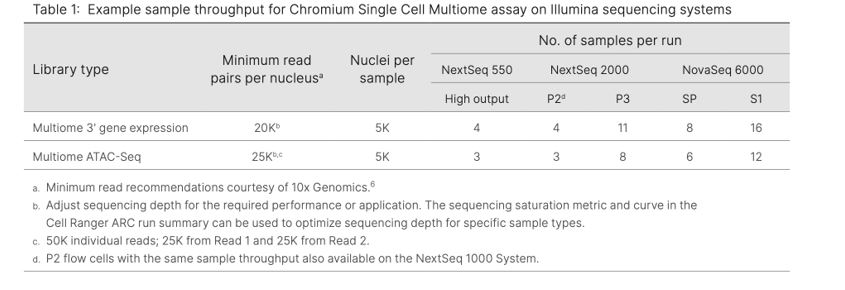
The 10x Genomics Multiome product is compatible with all Illumina® platforms, but we recommend sequencing on NextSeq™ 550/1000/2000 and NovaSeq™ 6000 to be able to sequence multiple samples per run (see table below). Due to differences in read configuration and recommended sequencing depths, Multiome gene expression libraries should be run on separate flow cells from Multiome ATAC libraries. Also note that NextSeq™ 550 requires a custom recipe, which can be obtained by contacting Illumina® Support (techsupport@illumina.com).
10x Genomics Multiome runs available on BSSH demo data page
To view a set of sequencing runs for 10x Genomics Multiome libraries run on both NextSeq™ 2000 P2 flow cells and NovaSeq™ 6000 SP v1.5 flow cells, please visit BaseSpace™ Sequence Hub. Check our previous blog post on our Demo Data section in BaseSpace Sequence Hub for additional details on how to access the run data on the demo data page.
Below are two links to directly import the runs and project folders into your account. These runs can be found under the category “Multiomics”. Because these are public data sets, these data sets are free and do not count against storage limits.
You can use the demo data run to compare with your own 10x Genomics Multiome runs. See our first blog post in this series on evaluating sequencing runs for additional details on how to estimate sequencing your run quality.
NextSeq™ 2000 P2 flow cells:
- Link to Multiome GEX run: https://basespace.illumina.com/s/ca148YJzl1lK
- Link to Multiome ATAC run: https://basespace.illumina.com/s/hey5s38fXQ7Y
- Link to Project folder with Cell Ranger ARC report files: https://basespace.illumina.com/s/mGueeP5OxjOm
NovaSeq™ 6000 SP v1.5 flow cells:
- Link to Multiome GEX run: https://basespace.illumina.com/s/91NBnxolGaqu
- Link to Multiome ATAC run: https://basespace.illumina.com/s/yHoG89YlB8Zi
- Link to Project folder with Cell Ranger ARC report files: https://basespace.illumina.com/s/XVLtuY4YLTvw
Access the 10x Genomics Cell Ranger ARC html report for the Multiome dataset
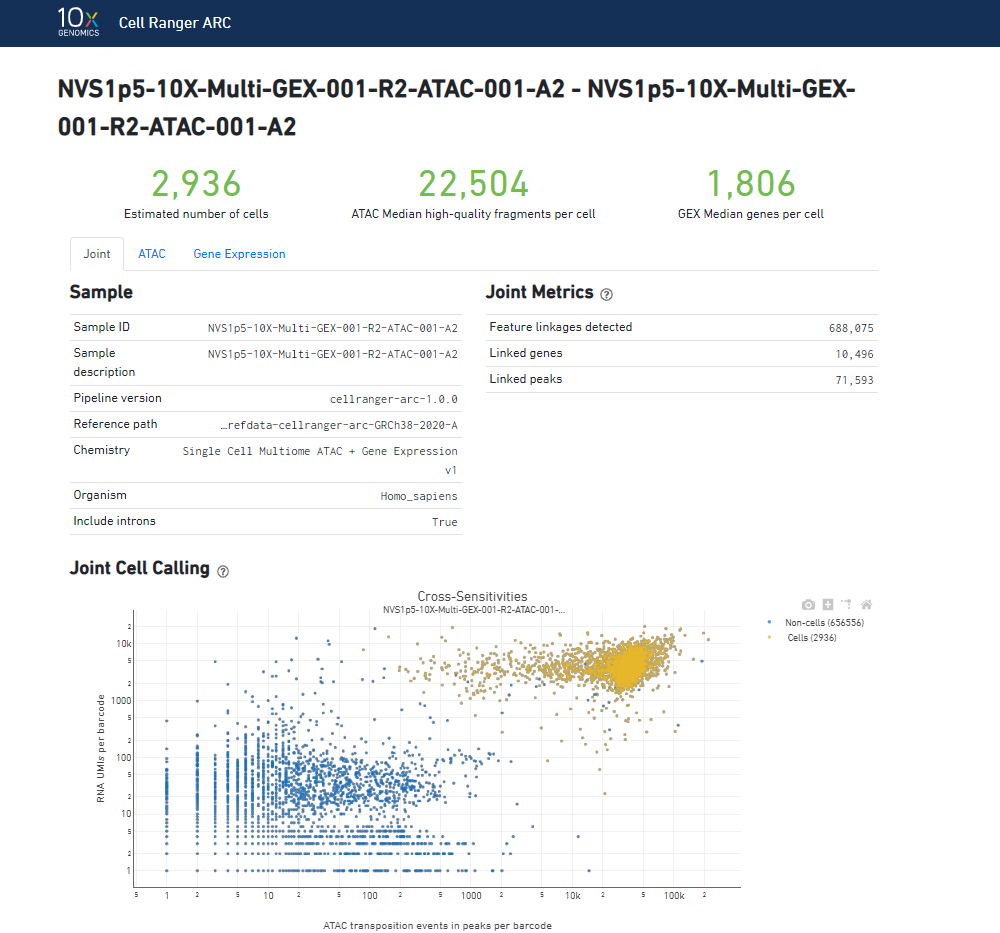
The html run reports from Cell Ranger ARC have been uploaded to the project, under the “Analyses” tab. 3 samples were provided by 10x Genomics and have been sequenced on these runs. The HTML report files generated by Cell Ranger ARC have been included in the project folder and can be downloaded from the “Files” tab.
The summary report contains summary metrics and automated secondary analysis results. The number of cells detected, ATAC median high-quality fragments per cell, and GEX median genes per cell are prominently displayed near the top of the page.
Additional resources
If you need further assistance in sequencing Chromium Single Cell Multiome ATAC + Gene Expression libraries, you can contact the Illumina® (techsupport@illumina.com) and 10x Genomics support teams (support@10xgenomics.com). The two teams collaborate to ensure you are fully supported throughout the workflow.
Additional resources and dataset examples are available here:
- Multiome App note: https://www.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/10x-multiomics-tech-note-m-amr-00006/10x-multiomics-tech-note-m-amr-00006.pdf
- Multiome product page: https://www.10xgenomics.com/products/single-cell-multiome-atac-plus-gene-expression
- Multiome software support page: https://support.10xgenomics.com/single-cell-multiome-atac-gex/software/pipelines/latest/output/summary
- Multiome datasets: https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets
Other posts from the blog series: new run data publicly available on BaseSpace™ Sequence Hub (BSSH).
Products mentioned here are for research use only, and are not for diagnostic use.
Special thanks to our colleagues at 10x Genomics for providing samples to sequence, and hat tip to Illumina Scientist Robin Bombardi Bombardi and the Emerging Applications Team for sequencing and analyzing these runs for this collaboration.