- Home
- News & Updates
- BaseSpace™ Sequence Hub now supports ScaleBio Single Cell RNA Sequencing Kit
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 11/03/2023
BaseSpace™ Sequence Hub now supports ScaleBio Single Cell RNA Sequencing Kit
We are delighted to present BaseSpace Sequence Hub data for the ScaleBio Single Cell RNA Sequencing Kit.
Many single-cell capture systems face throughput restrictions imposed by the need to physically sequester single cells in unique compartments. The ScaleBio Single Cell RNA Sequencing Kit unleashes throughput by using combinatorial indexing and an instrument-free approach to achieve highly multiplexed library preparation on hundreds of thousands of cells at a reduced overall cost relative to existing workflows.
Workflow
Using the cell as the reaction compartment cell transcripts are taken through 3 sequential rounds of barcoding, performed by distributing on an indexing plate, pooling, and redistributing for two additional rounds of barcoding. This three-level split-pool approach results in 3.5 million barcode combinations, enabling the profiling of hundreds of thousands of cells while maintaining a low multiplet rate (<5%) and reducing running costs. For more details on the protocol, please visit the ScaleBio Support Page.
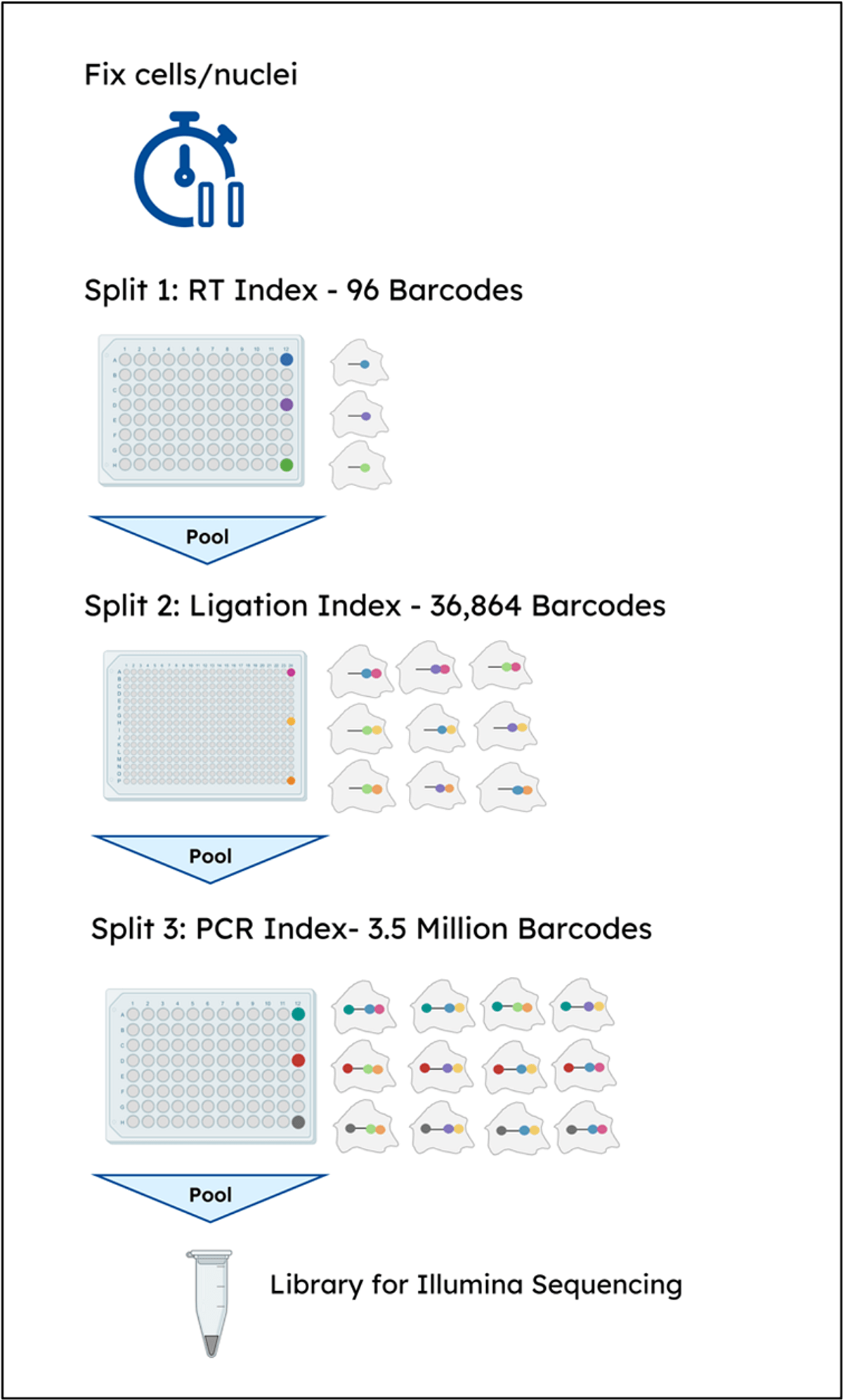
Figure 1. Library preparation workflow using the ScaleBio Single Cell RNA Sequencing Kit.
Once all indexes have been incorporated, the final library is made by pooling material from the desired number of wells for cleanup and sequencing. As each well of the Final Distribution Plate contains a small but representative subsampling of all samples that were loaded into the first (RT) plate, it is possible to sequence the entire library (an output of ~125,000 cells) or perform deep sequencing on a small subset of cells for QC before committing to a larger sequencing run on a mid-throughput instrument such as the NextSeqTM 2000. The target depth is ~20,000 reads/cell for standard single-cell phenotyping.
Sequencing Run on NextSeq 2000 Illumina platform
Below are two links to directly import the runs and project folders into your BaseSpace account. These runs can be found under the “single cell” and “RNA-seq” category. Because these are public data sets, these are free and do not count against storage limits. See our blogpost to understand how to access these data.
You can use the demo data to compare with your own single cell runs. See our first blog post in this series for additional details on how to evaluate your sequencing run quality.
Run link https://basespace.illumina.com/s/Jv0GHVIB4c3r
Project link: https://basespace.illumina.com/s/8Og9vQpte3Y0
In this data set, PBMCs were loaded at 10,000 cells per well, for a total of 960,000 cells across the RT Barcode Plate. A total of 12 wells from the final library were sequenced on a NextSeq 2000 P2 flowcell. The loading concentration of the pool was 650 pM. No PhiX was spiked in this run. Illumina always recommends spiking some PhiX as a positive control for the sequencing run.
The recommended read length for the Scale single RNA-seq library is 34 + 10 + 76. With Q30 = 88.23% and a yield of 49.05 Gbp, this run is meeting Illumina specifications for Q30 and yield for a P2 flowcell (https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000/specifications.html).
Access the analysis report on BaseSpace Sequence Hub
The html analysis reports have been uploaded to the project, under the “Analyses” tab. They have been included in the project folder and can be downloaded from the “Files” tab.
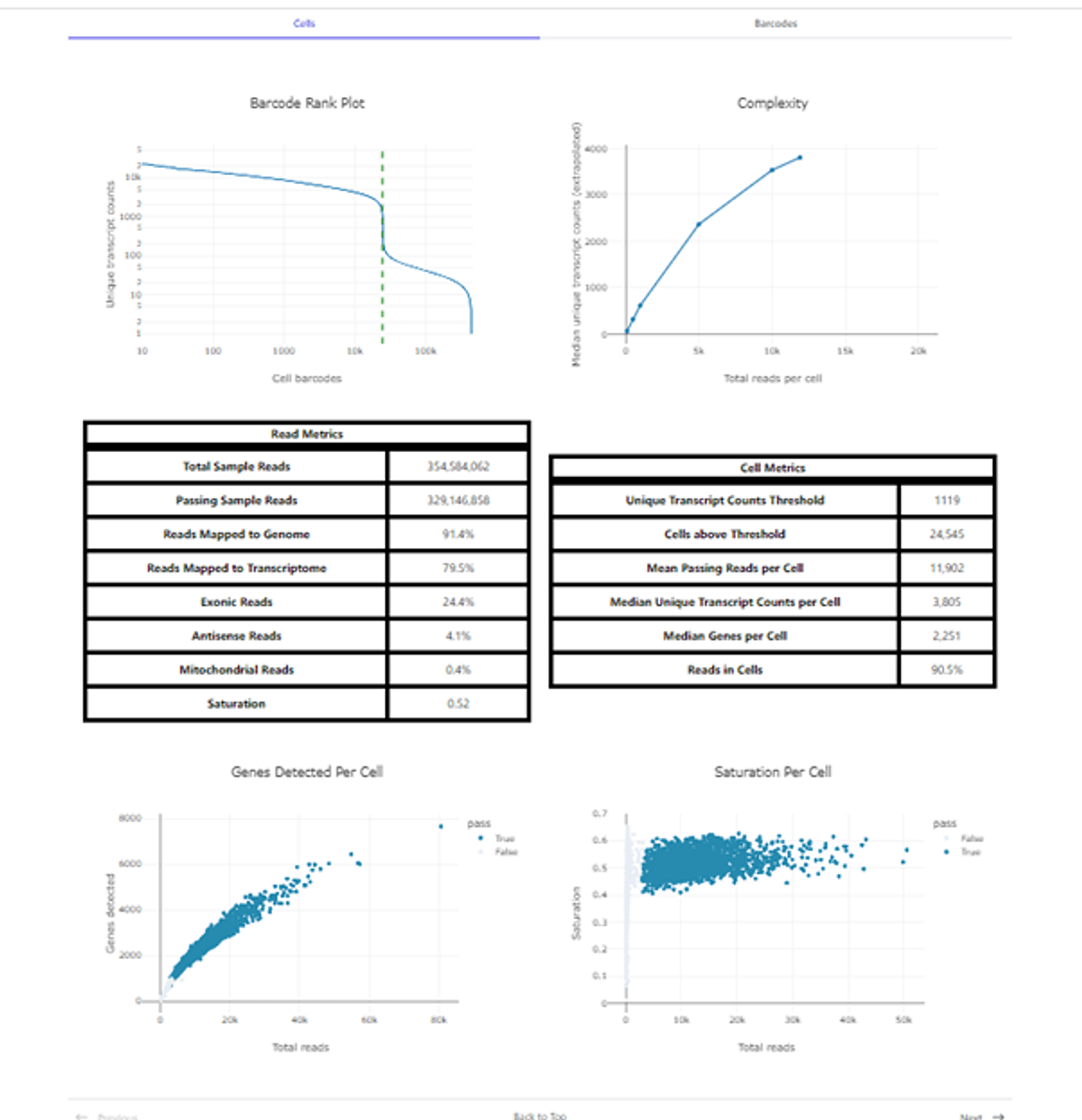
Figure 2: Scale single cell RNA-seq analysis report example.
In this data set, PBMCs were loaded at 10,000 cells per well, for a total of 960,000 cells across the RT Barcode Plate. A total of 12 wells from the final library were sequenced on a NextSeq 2000, and 24,545 cells were recovered. Extrapolated to the full library (pooling material from all 96 wells) this would equate to a recovery of ~196,000 cells in total while maintaining a low multiple rate of ~5.5%. Alongside the high throughput, the ScaleBio Single Cell RNA Sequencing Kit yielded high quality data, with ~80% of reads mapping to the transcriptome, >90% of reads found in cells, and 3,805 median transcripts per cell detected across 2,251 genes (50% saturation). Together these data demonstrate Scale Biosciences’ highly efficient chemistry for construction of complex and high sensitivity single-cell libraries for high-throughput single-cell profiling.
Support
Contact Scale Biosciences for any questions regarding the single cell library prep assay and analysis. Contact Illumina Tech support for sequencing related questions.
Special thanks to our colleagues at Scale Biosciences for providing the data.
For Research Use Only. Not for use in diagnostic procedures. M-GL-02329