- Home
- News & Updates
- BD Rhapsody TCR/BCR Profiling Assay data on the NextSeq™ 2000 – 600 cycles kit are now available on Illumina BaseSpace™ Sequence Hub!
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 09/27/2023
BD Rhapsody TCR/BCR Profiling Assay data on the NextSeq™ 2000 – 600 cycles kit are now available on Illumina BaseSpace™ Sequence Hub!
Today, we are excited to present that NextSeq 2000 data on the newly launched 600 cycles sequencing kits with the BD Rhapsody TCR/BCR Profiling Assay are now available on BaseSpace Sequence Hub.
Analyzing paired chain information of T cell receptors (TCR) and B cell receptors (BCR) at the single-cell level is a powerful tool for probing T and B cell diversity and function in lymphoid malignancies, infectious diseases, autoimmune disorders and tumor immunology. The BD Biosciences TCR and BCR profiling tools help uncover clonal diversity and function of T and B cells, respectively, at the single-cell level. The BD Rhapsody TCR/BCR Profiling Assay helps assemble the full length VDJ sequence for the T cell and B cell receptor while assessing transcriptomic and protein expression of the same cells.
Sample, library, and sequencing preparation
The library pool was spiked with 20% PhiX and diluted to 650pM prior to sequencing with the NextSeq 1000/2000 P2 Reagents (600 Cycles) catalog number 20046813.
Accessing sequencing run
To view this run on NextSeq 2000 600 cycles kit, please access the BaseSpace Sequence Hub and import the sequencing run and FASTQ files information using the links provided below. These runs can be found under the category “Immune Repertoire Sequencing” and “Multiomics”. Because these are public data sets, these data sets are free and do not count against storage limits.
Run link: : https://basespace.illumina.com/s/DhSpmR20RUjK
Project link: https://basespace.illumina.com/s/XezdXtJ1gfQE
Check blog post "Demo Data Page" for additional details on how to access the run data on the demo data page.
Evaluation of the sequencing run
You can use the demo data run to compare with your own BD Rhapsody TCR/BCR Profiling Assay sequencing runs. Check the blog post “Does my sequencing run look good” for additional details on how to estimate your sequencing run quality.
With a Q30=77.58%, this run is just below the 80% specification for the 600 cycles kit (https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000/specifications.html). However, please note that quality scores are based on NextSeq Reagent Kits run on the NextSeq 1000 and 2000 system using an Illumina PhiX control library, which is a well balanced library. Performance may vary based on library type and BD Rhapsody libraries include several regions of very low base diversity.
With a yield of 226 Gb, this run exceeds the specifications (180 Gb for the NextSeq 2000 P2 600 cycles kit).
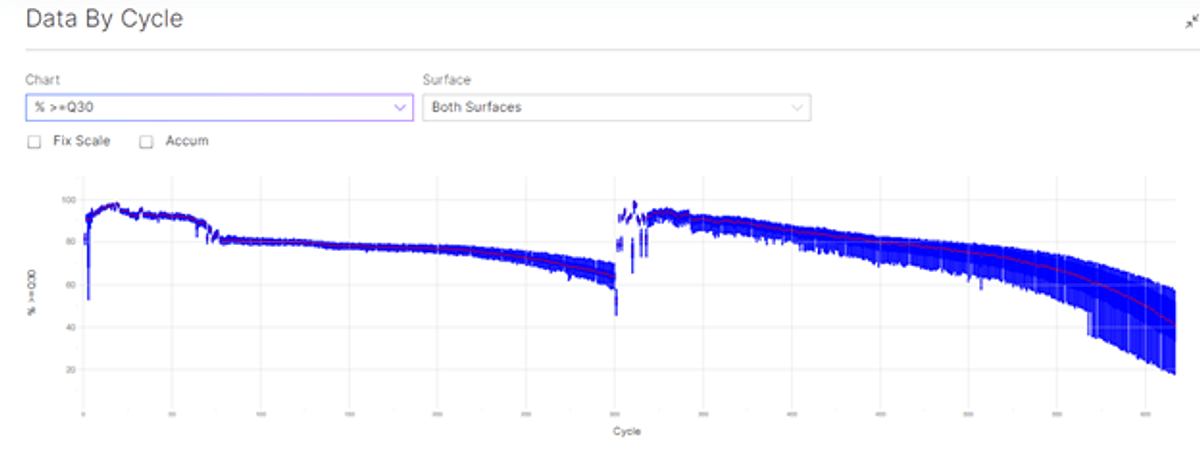
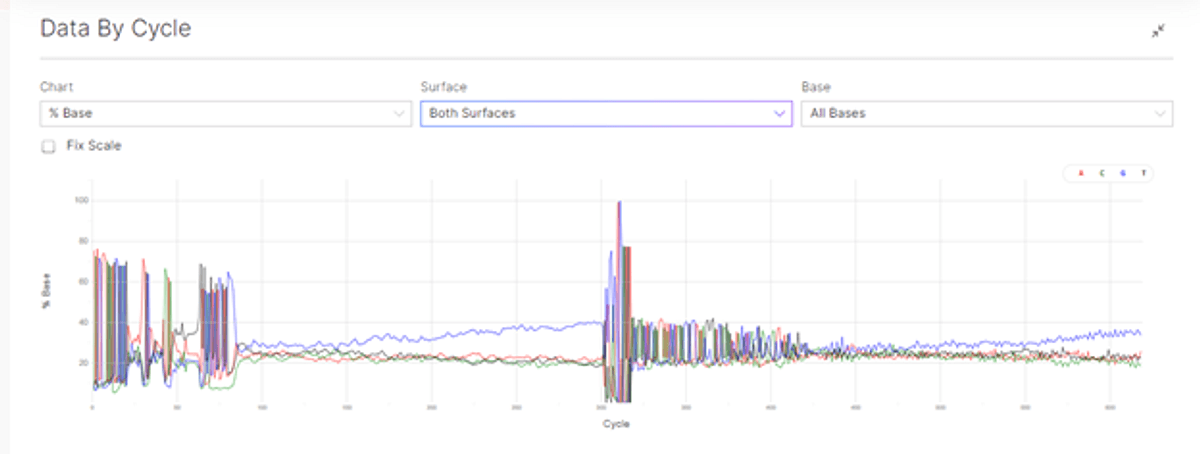
Figure 1: sequencing data by cycle showing the data by cycle for Q30 (top), and percentage base by cycle (bottom).
Analysis
Sequencing data were analyzed with BD Rhapsody Sequence Analysis Pipeline version 2.0. The HTML report files generated by the pipeline have been included in the project folder and can be downloaded from the “Files” tab (under the “Report html” folder).
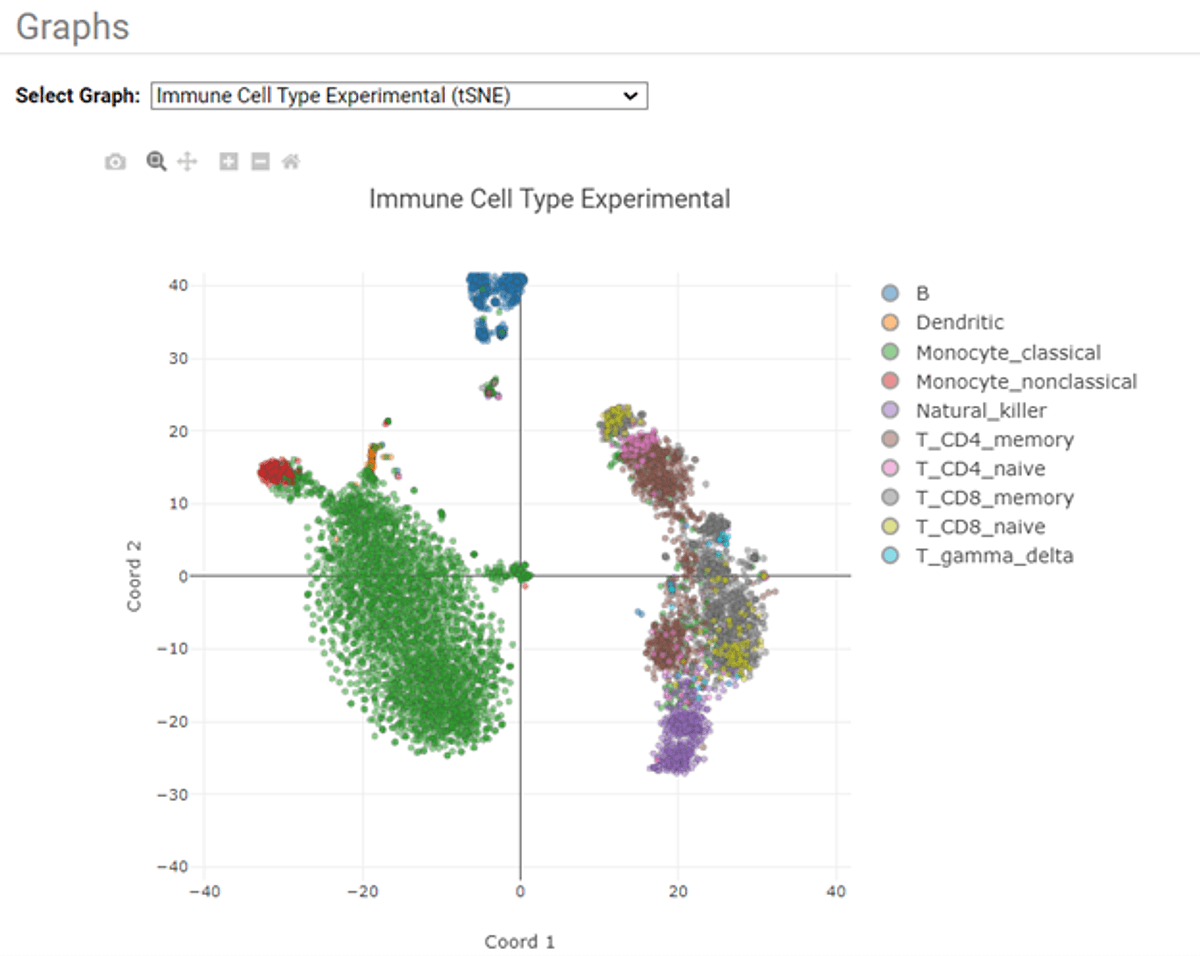
Figure 2: t-SNE plot generated by the BD Rhapsody Sequence Analysis Pipeline version 2.0.
Support
Contact BD for any questions regarding the single cell library prep assay and analysis. Contact Illumina Tech support for sequencing related questions.
Additional resources:
https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000/specifications.html
https://www.bdbiosciences.com/en-us/products/reagents/single-cell-multiomics/vdj#Overview
Special thanks to our colleagues at BD Biosciences for providing all the libraries. Hat tip to Illumina Scientists Robin Bombardi and Qiyuan Han and the Emerging Apps team for sequencing these runs for this collaboration.
For Research Use Only. Not for use in diagnostic procedures M-GL-02130