- Home
- News & Updates
- Connected Insights v5.1: Improved visualizations, heme interpretation, and streamlined curation capabilities
-
Illumina Connected Insights
-
Product updates
-
News
- 02/12/2025
Connected Insights v5.1: Improved visualizations, heme interpretation, and streamlined curation capabilities
The Illumina Software and Informatics team has been hard at work over the past year, continuously building on recent innovation within the Connected Software portfolio. Latest updates provide our customers with breakthrough solutions for the most accurate and efficient analysis and interpretation possible.
Last month, Illumina released Connected Insights v5.1, providing oncology customers with new variant curation capabilities that make using our flagship oncology interpretation platform even easier to use. Building on version 5.0 and earlier releases, the latest iteration of Connected Insights makes clinical research variant interpretation more powerful for a range of oncology use cases such as heme malignancy clinical research.
Features such as improved visualizations, a new heme interpretation toolkit, multi-version support (for software version updates to fit on your schedule), and additional curation capabilities (such as the ability to add a virtual variant, one-letter p.HGVS notations, harmonization of tiering levels across knowledge sources) are all included in the release of Connected Insights v5.1 to empower our customers with operational efficiency and streamlined insights.
Connected Insights v5.1 highlights include:
- Improved visualization functionality, with addition of BAM tracks to biomarker visualization plots and integration of desktop IGV
- Direct integration with latest DRAGEN pipelines for heme (DRAGEN Heme WGS and DRAGEN Amplicon) and new AML subtype auto-classification tool for heme interpretation – learn more here specific capabilities released here.
- Multi-version support, providing customers with control when to upgrade to the latest version released
- Enhanced curation and reporting capabilities including the ability to add a virtual variant, support for one-letter hgvsp and harmonization of KB specific tiers to a standard
Let’s jump into a few of the details behind the Connected Insights v5.1 release:
Improved visualizations
As part of our latest iteration of Connected Insights, version 5.1 integrates several improvements to existing visualization functionality. DNA and RNA BAM tracks have now been added to the visualization tab, further advancing functionality for variant QC and interpretation. For example, when exploring breakpoints and protein domains on fusion plots, users can evaluate reads supporting the fusion calls in the same view, streamlining the process. Additionally, the desktop version of IGV has now been integrated, making full IGV functionality accessible to users.
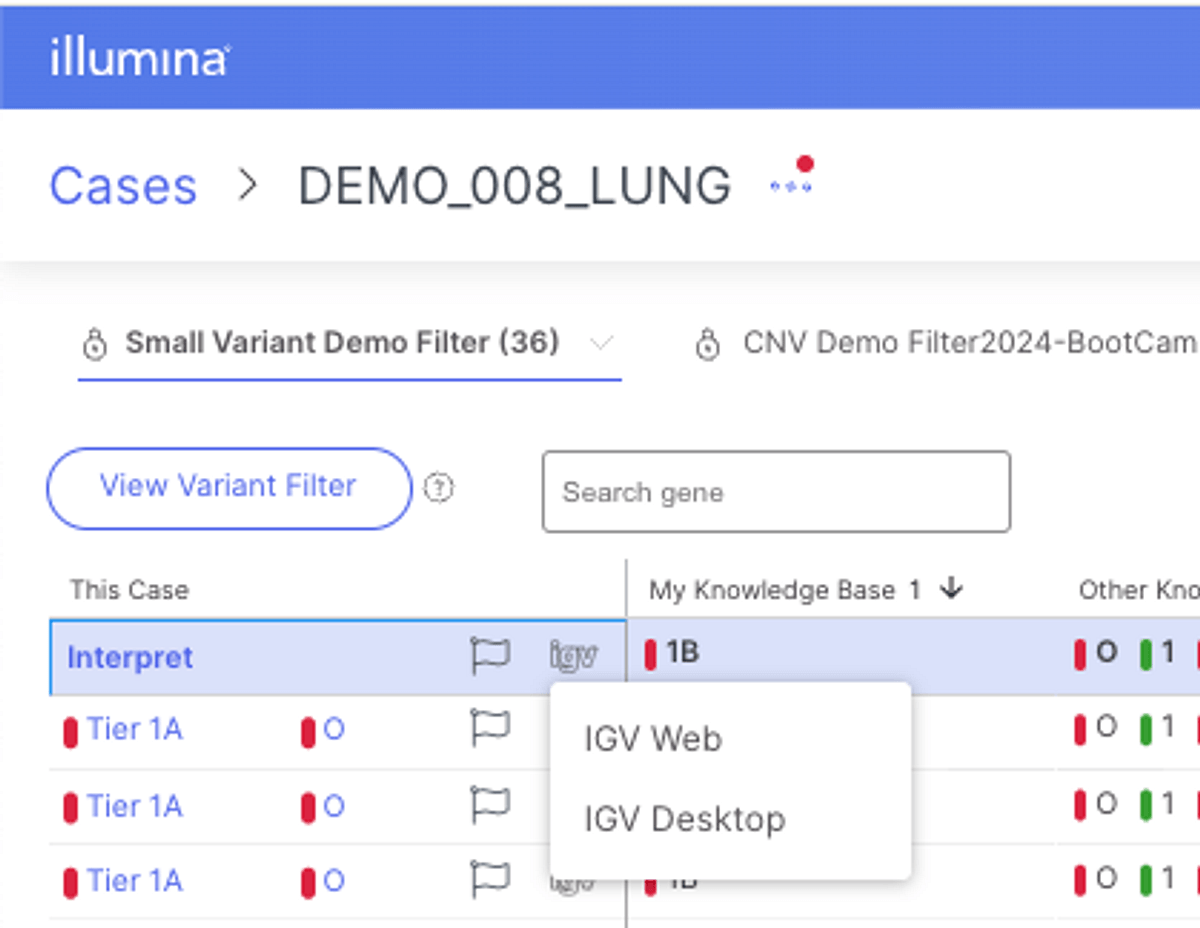
Curation and Reporting Capabilities: Virtual variants, harmonization of KB tiers, one-letter p.HGVS notations
Connected Insights integrates over 55 powerful knowledge sources, which can be leveraged to curate and interpret variants before adding to a research report. As tiering levels can vary across knowledge bases, Connected Insights now offers the capability of harmonizing tiering in reports based on ASCO/AMP, ESCAT, or custom guidelines. New auto conversion functionality makes it simpler and easier to understand reports while maintaining compliance with reporting guideline recommendations.
We know that many well-known variants within oncology are referred to using one-letter HGVS notations, a personal preference of many. Connected Insights v5.1 makes finding variants faster, enabling our users to filter and select based on one-letter or three letter p.HGVS notations – both of which can be displayed side by side.
Connected Insights also now allows users to manually add variants not present in the VCF or not called from FASTQ to their case. This is useful when needing to complement NGS data with data ingested from other genetic tests (long-read sequencing, optical mapping, CGH, SNP array, karyotyping/FISH, repeat-primed PCR, MLPA, Southern blot, etc).

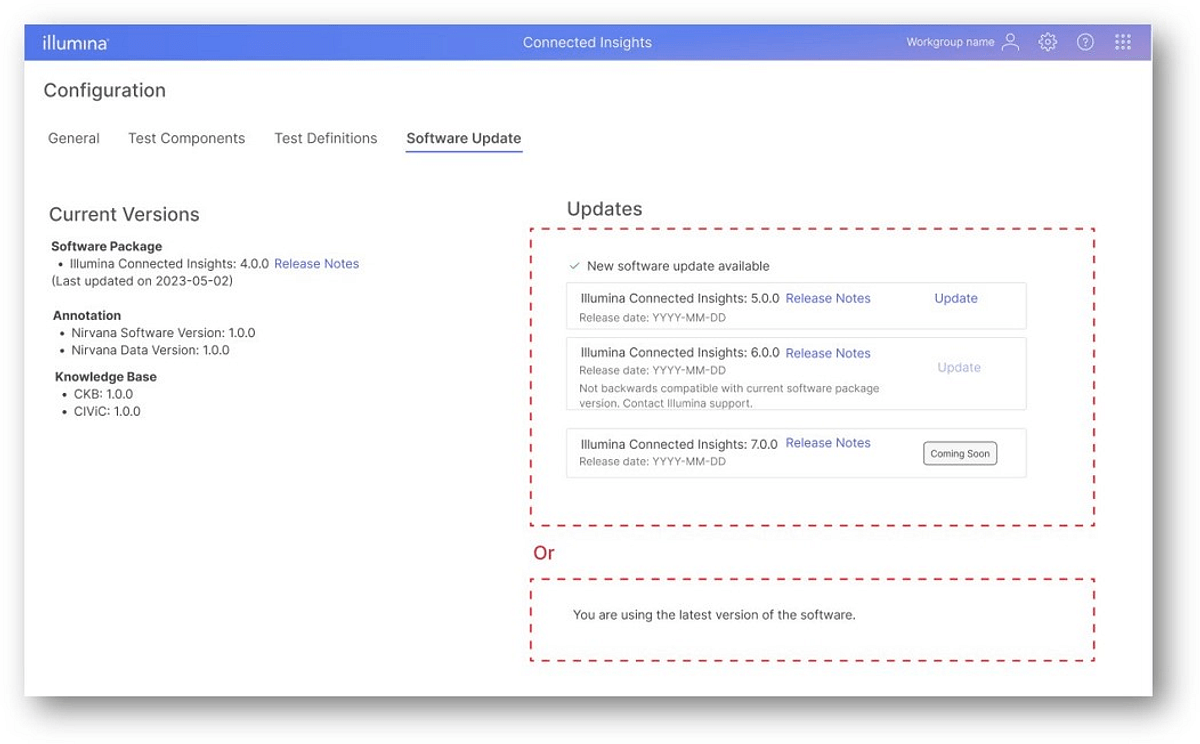
Upgrades on demand with multi-version support
Building on functionality first released in v5.0, Connected Insights now enables users with full control of when to upgrade the software. Users are notified within the platform before and after deployment of a new version, and can initiate an upgrade to the newly available versions in the Software Update page on their own schedule. Customers can choose to upgrade one, several, or all workgroups as they see best fit. Beginning with version 5.1, Connected Insights will support versions for 2 years after a release, and any End of Life (EOL) notifications will be delivered 12 months prior to the date.
Other key features
In addition to the features mentioned, heme interpretation capabilities are at the forefront of Connected Insights version 5.1. To learn more about heme-specific capabilities included in the latest version released, click here.
Learn More
Visit the Connected Insights webpage to learn more about our technology and speak with a Technical Specialist. If you are interested in connecting with us and the growing community of users, please also let us know!
For Research Use Only. Not for use in diagnostic procedures.
M-GL-03437