- Home
- News & Updates
- Fully featured Heme analysis: DRAGEN secondary analysis and Connected Insights v5.1 heme interpretation capabilities
-
Illumina Connected Insights
-
Product updates
-
News
- 02/12/2025
Fully featured Heme analysis: DRAGEN secondary analysis and Connected Insights v5.1 heme interpretation capabilities

Hematological malignancies represent an important field within cancer genomics research, helping researchers better understand the molecular drivers and etiology behind blood cancers. While Illumina has supported heme malignancy research for more than a decade, new breakthrough innovations within Illumina’s informatics solutions for oncology, DRAGEN secondary analysis and Illumina Connected Insights, now drive more powerful, comprehensive interpretation of heme NGS data than ever.
Hematologic malignancies account for a large number of new cancer cases diagnosed in the United States each year, at around 9.5%. Characterization of genomic mutations at the many different stages of hematopoietic differentiation can help better understand the molecular drivers behind distinct tumor subtypes and disease etiology of blood cancers, unlocking new insights into myeloma, lymphoma, and leukemia-associated genes. In contrast to traditional single-gene methods, targeted and whole genome next-generation sequencing (NGS) offers significant advancements in sensitivity and scale, providing greater visibility into important drivers of hematologic malignancies.
While Illumina has provided long-standing support for NGS hematological samples through targeted sequencing panels (AmpliSeq Myeloid) and WGS (Illumina DNA PCR-Free Prep), variant calling and interpretation of heme WGS NGS data has remained a major bottleneck for researchers until recently. With a lack of interpretation software, deriving new insights proved to be incredibly challenging. It requires users to the manually combine of results from multiple technologies (FISH, FCR) and apply complex guidelines with multiple parameters to determine disease subtype and risk level.
Recent advancements within Illumina’s oncology informatics portfolio are now making powerful analysis and interpretation of NGS heme data available for the first time ever. Illumina now offers fully featured interpretation capabilities through recent releases made to its secondary analysis and interpretation solutions for oncology: DRAGEN secondary analysis and Illumina Connected Insights for variant interpretation. The release of DRAGEN v4.3 in June 2024 combined with the most recent launch of Connected Insights v5.1 make a combined, single-vendor solution for heme malignancy clinical research accessible for the first time ever. Latest features released within Connected Insights v5.1 including powerful visualizations such as Circos and other plots, AML subtype auto-classification, and integration of non-NGS data (karyotypes, blast count %) provide researchers with a comprehensive, powerful toolkit to visualize and interpret data from hematological malignancies.
Highlights of Connected Insights heme capabilities include:
- Direct integration of latest DRAGEN pipelines for heme within Connected Insights, including DRAGEN Heme WGS and DRAGEN Amplicon
- Powerful visualization functionality including interactive Circos plot and enhanced fusion plots, providing alternative views to linear genome view and enabling the exploring of large and complex biological events (SVs, CNVs) in addition to IGV desktop integration
- Access to 55+ external knowledge sources, with heme-specific content from guideline and clinical trials from CKB, OncoKB and more
- AML subtype auto-classification based on WHO 2022 & ELN 2022 guidelines and access to an interactive evidence map
Let’s jump into the details of a few of the new heme interpretation capabilities now embedded within Connected Insights version 5.1:
Integration of latest DRAGEN Heme Pipelines
Comprehensive genomic analysis of hematologic malignancies requires the evaluation of multiple variant types, including small variants (single-nucleotide variants and insertions-deletions smaller than 50 base pairs), copy number alterations (CNAs), structural variants (SVs) such as translocations, and loss of heterozygosity (LoH) calculation. Connected Insights v5.1 introduces updated compatibility with state-of-the-art DRAGEN heme pipelines, including DRAGEN Heme WGS and DRAGEN Amplicon. Using a combination of variant calling, depth of coverage analysis, break-end detection, and other algorithms, the DRAGEN Somatic WGS Heme Tumor Only recipe provides accurate and comprehensive results for variants associated with hematologic malignancies. DRAGEN Heme WGS analysis pipelines, made available for early access in DRAGEN v4.3 and general access in v4.4, enables key heme analysis features including:
- SNPs & INDELs: Best in class variant calling for SNPs and INDELs in genes such as NPM1
- Internal Tandem Duplications: Increased sensitivity for ITDs within genes such as FLT3, KMT2A.
- SVs: Large structural variants detection including recurrent gene fusions such as PLM::RARA and BCR::ABL1
- DUX4 Rearrangements: Detection of rearrangements in difficult to map regions, such as DUX4::IGH
- Purity/Ploidy Aware: Tumor purity estimation enables accurate calling of chromosomal ploidy states
- LOH: Allele specific copy number and LOH calling
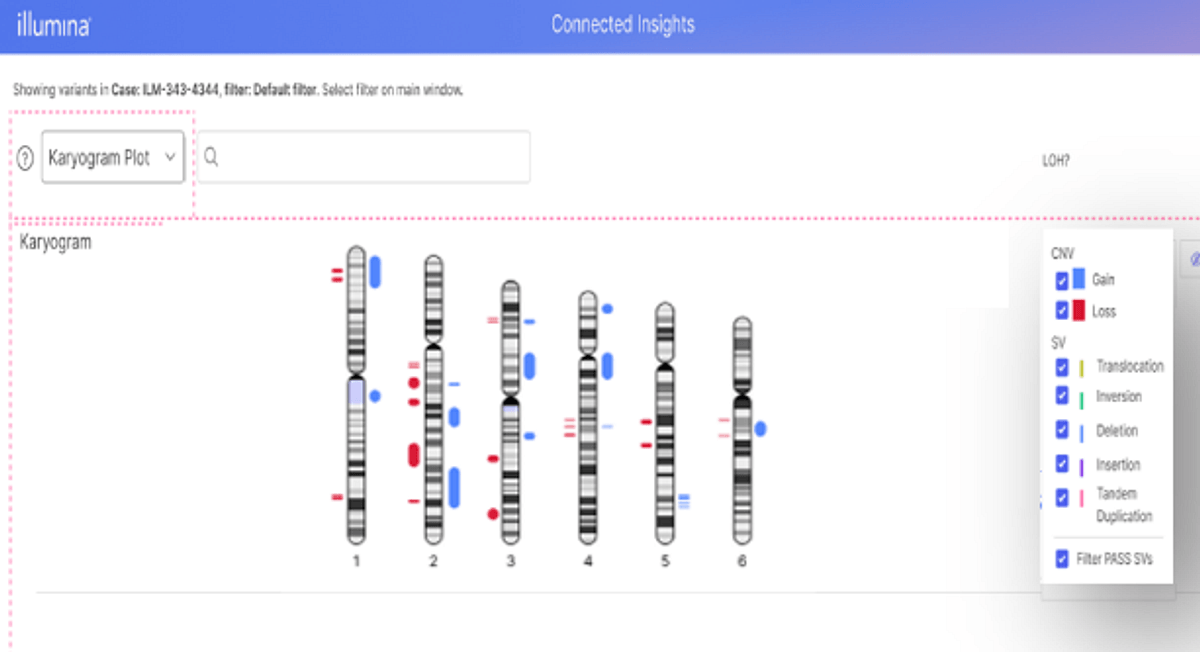
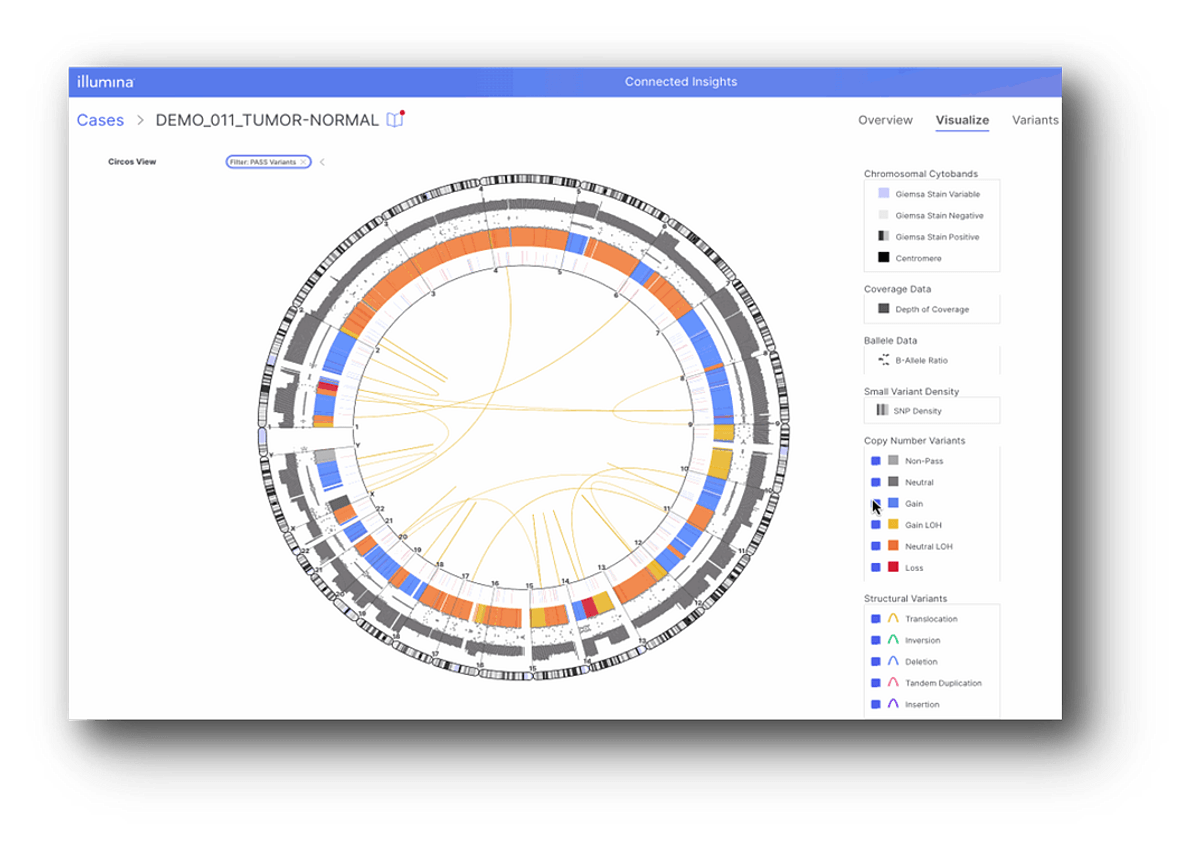
Powerful heme visualizations: Linear Genome View, Fusion and Circos Plots
Over the past few releases, Connected Insights has built out a powerful toolkit of visualization capabilities providing a one-stop-shop for hematological malignancies, including linear genome views and Circos Plots.
Circos plots are a commonly used visualization tool in the field of comparative genomics to map and displayed variation in genomic data across chromosomes, all within a single view. Connected Insights also integrates powerful linear genome views to make visualizing complex heme WGS data easier to see.
The latest release of Connected Insights now additionally offers improvements to visualizations of fusions, bringing integration of IGV desktop (previously solely available on cloud) to enable an even more robust heme interpretation toolkit.
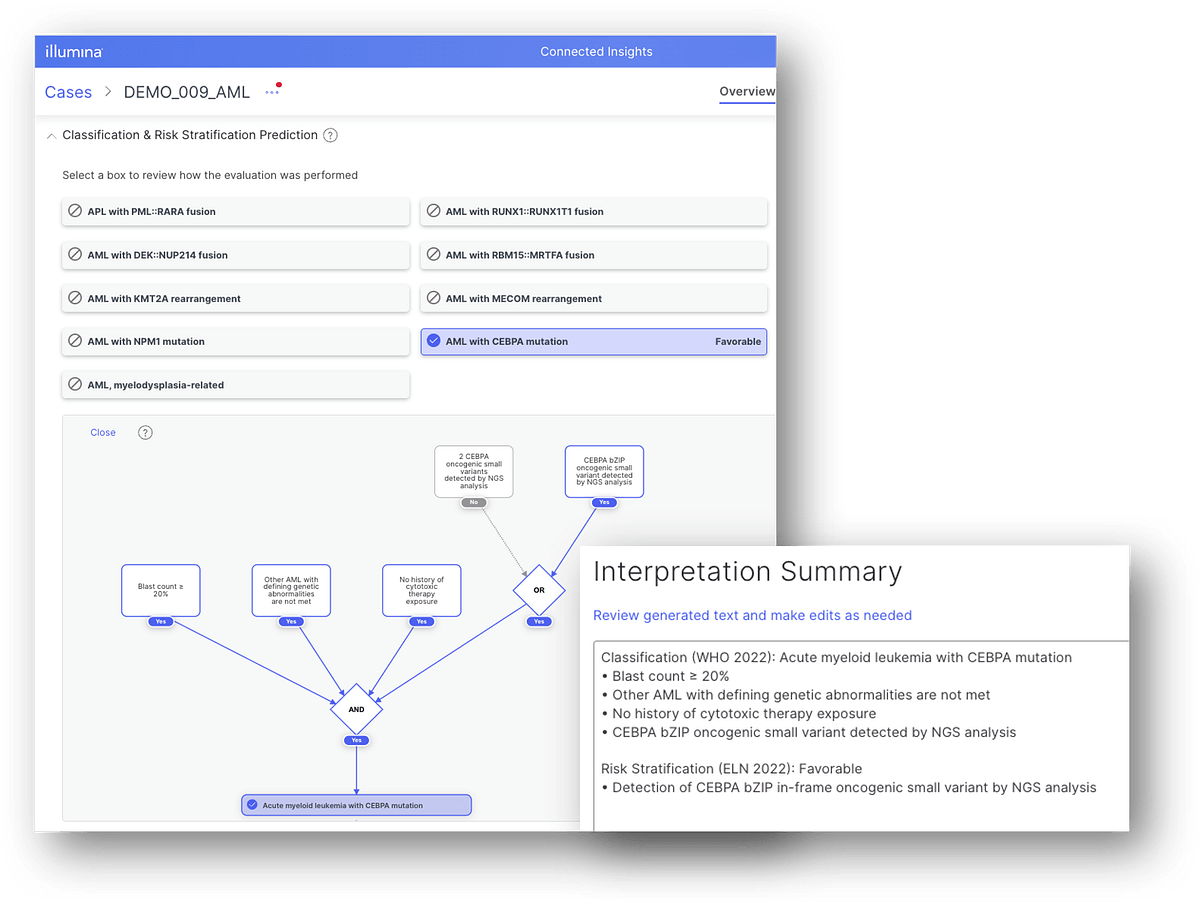
Enhanced curation capabilities with AML subtype auto-classification
Connected Insights v4.0 and v5.0 introduced new functionality for automating guidelines-based oncogenicity classification, incorporating latest and greatest Illumina PrimateAI-3D and SpliceAI algorithms as part of its oncogenicity prediction scoring framework. Now as of v5.1, Connected Insights leverages the same set of algorithms to determine AML subtype and risk based on biomarkers and other data. Based on WHO 2022 and ELN 2022 risk stratification guidelines, users can ingest NGS (targeted panels, WGS) or non-NGS (eg, karyotype, blast count %) heme data and automatically classify AML subtypes, with access to an interactive evidence map and decision tree to verify accuracy of results.
Other key features
While heme interpretation capabilities were at the forefront of features released in version 5.1 of Connected Insights, we’ve only touched the surface when it comes to other features released.
Additional Connected Insights v5.1 features include:
- Multi-version support upgrade process – Control when to upgrade to the latest version released, finding available versions in the Software Update page and initiate the upgrade in one/several workgroups as desired
- Manual data upload from BaseSpace Sequence Hub for legacy Amplicon pipelines– In addition to automated case creation and VCF data ingestion, existing BaseSpace Sequence Hub users can now create cases manually as needed by browsing to existing analysis results from legacy Amplicon pipelines
- One-letter hgvsp– Set personal preference to using one-letter or here-letter HGVS as an easier, faster way to find variants
- Auto-conversion of KB specific tiers to a standard – harmonize tiering in reports based on ASCO/AMP, ESCAT, or custom guidelines to make simpler, easier to understand reports while maintaining compliance with reporting guideline recommendations
Learn More
Visit the Connected Insights webpage to learn more about our technology and speak with a Technical Specialist. If you are interested in connecting with us and the growing community of users, please also let us know!
For Research Use Only. Not for use in diagnostic procedures.
M-GL-03375