- Home
- News & Updates
- DRAGEN™ 4.2: Enhanced machine learning, new targeted callers, and more
-
DRAGEN
-
Product updates
- 07/11/2023
DRAGEN™ 4.2: Enhanced machine learning, new targeted callers, and more

We’re celebrating 5 years of DRAGEN at Illumina with DRAGEN version 4.2! Version 4.1 enabled high throughput DRAGEN workflows on NovaSeq X, now v4.2 continues to push the boundaries of accuracy and comprehensiveness for secondary analysis on cloud and on-premises. Access the latest software and learn more about new features on our support page.
May 2023 marked 5 years of DRAGEN at Illumina and over 25 years of Illumina innovation. We are thrilled to celebrate these anniversaries by releasing our most accurate, most comprehensive version of DRAGEN secondary analysis ever. DRAGEN v4.2 continues to expand its award-winning accuracy1,2 with major Germline accuracy improvements driven by updated references and improved machine learning. Additionally, this version introduces a lineup of exciting new features. In this blog, we will delve into significant small CNV accuracy improvements, expanded caller offerings, greater gVCF Genotyper efficiency, and much more.
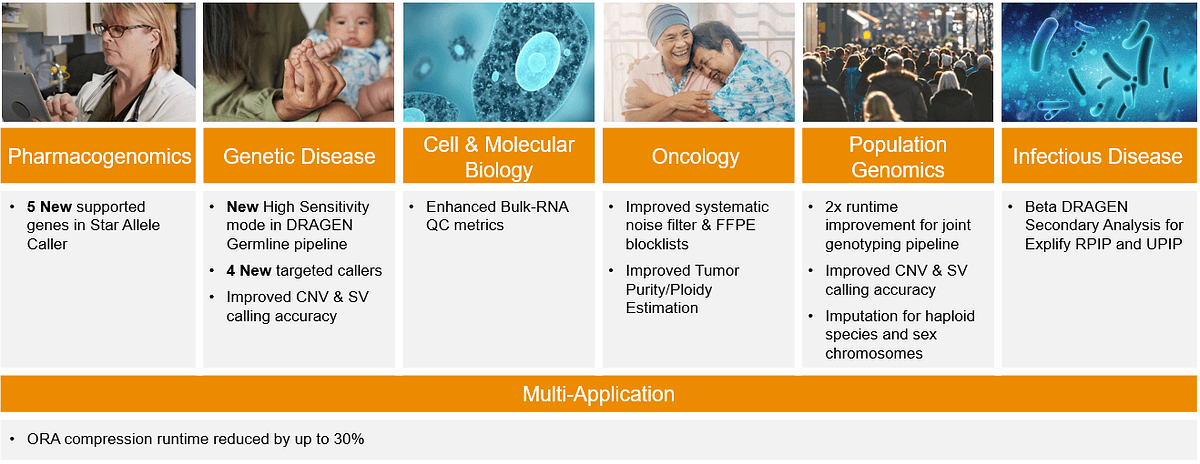
As DRAGEN continues to serve and support comprehensive application areas, here is a quick preview of the updated key features in DRAGEN v4.2 before we take a closer look at a few major improvements.
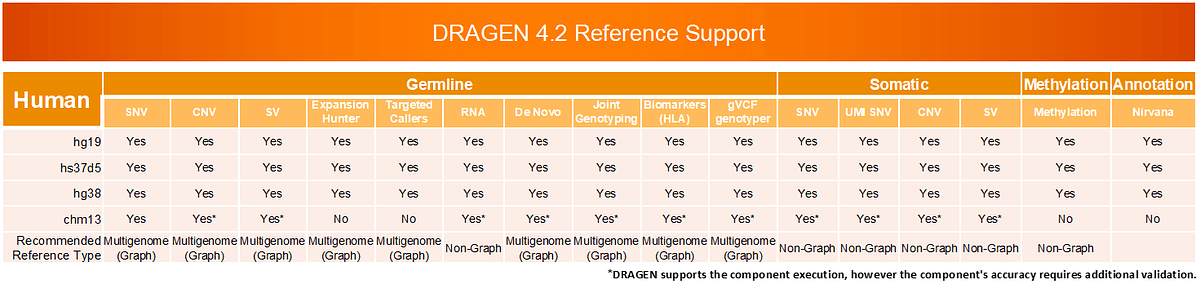
Germline Accuracy Improvements
We’ve made multiple updates to the reference genomes to drive significant accuracy gains.
- New, more diverse population haplotypes integrated with the DRAGEN Multigenome (graph) reduce non-European small variant calling errors by 15-17%.
- With the updated hg38 reference and Multigenome (graph) enabled, DRAGEN v4.2 reduces errors of small variant calling in Challenging, Medically Relevant Genes (CMRGs) by 25% compared to DRAGEN 4.0.
- DRAGEN v4.2 now supports the latest Telomere-to-Telomere CHM13v2.0, the first complete gapless sequence of a human genome, that enables improved small variant calling.
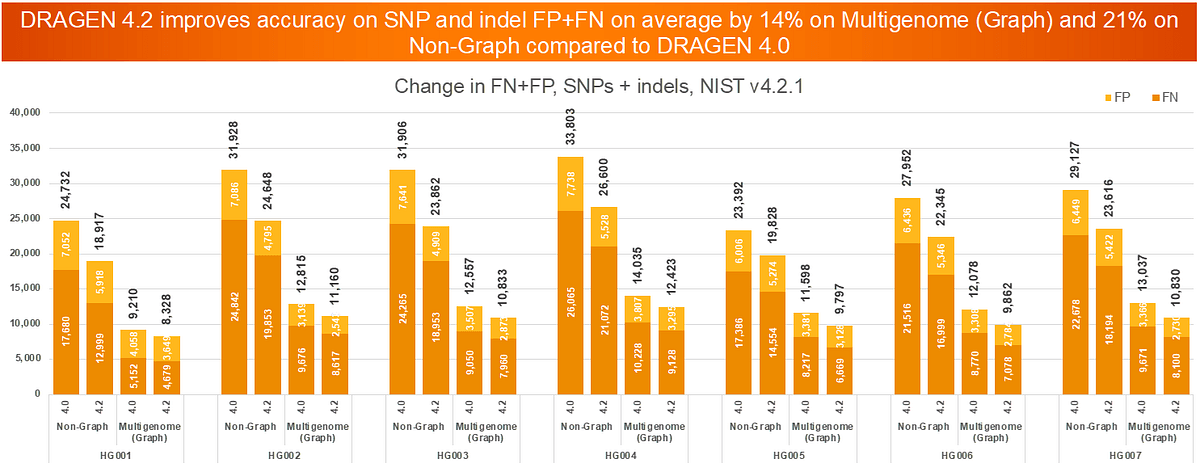
Updated DRAGEN-ML and Multigenome (Graph) enables an average SNV error reduction of 14%.
Starting from DRAGEN v4.0, the default setting includes DRAGEN-ML, which reduces SNV errors. We recommend enabling Multigenome (Graph) in your analysis to further contribute to significant error reduction.
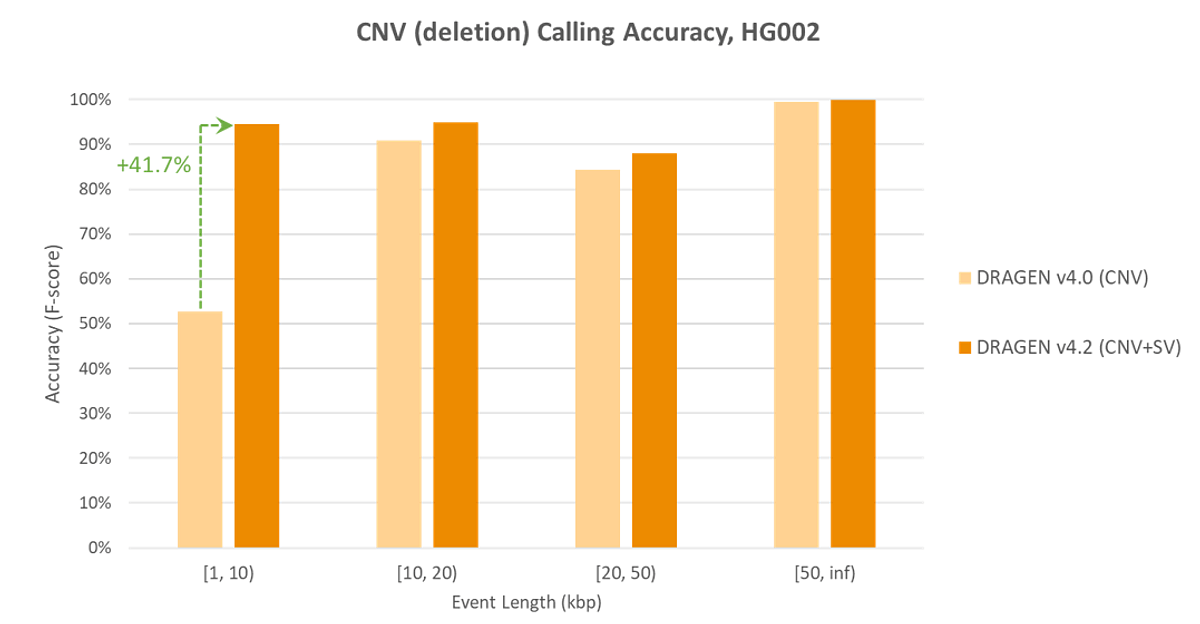
Significant Accuracy Improvements for Small CNV
Increase Copy Number Variant (CNV) accuracy in DRAGEN v4.2 with CNV detection that leverages Structural variation (SV) information.
New joint SV and CNV caller enables higher accuracy at all event size. Most notably, gains of over 90% in recall, 25% in precision, and 41.7% in the F1 score are achieved for CNVs in the 1kbp-10Kbp range.
Expanded Caller Offerings
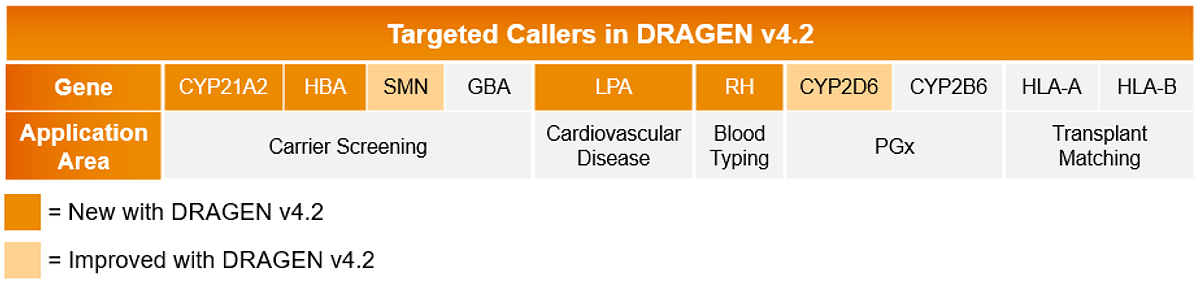
Increase accuracy in focused variant detection with 4 new targeted callers.
Improve carrier screening insights with new targeted callers CYP21A2 and HBA. Expand cardiovascular and blood typing insights with the new targeted callers LPA and RH, respectively. Improving upon previous callers, DRAGEN v4.2 also improves CYP2D6 calling accuracy and supports SMN silent carrier calling.
Learn more about our recent callers:
- Overcoming high homology to detect variation in CYP21A2 with whole-genome sequencing in DRAGEN.
- Genotyping of high homology HBA1 and HBA2 from Illumina whole-genome sequencing.
- Using whole-genome sequencing to evaluate copy number variants of the LPA Kringle-IV type 2 domain with DRAGEN.
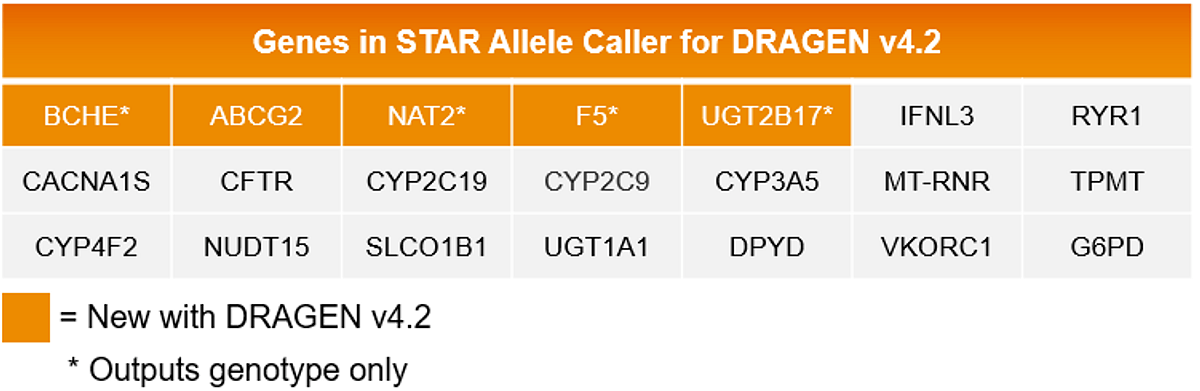
Improve pharmacogenomic insights into optimal genotype and corresponding metabolism status with 5 new genes in STAR allele caller.
A More Efficient gVCF Genotyper
Produce more insights, in half the time with the improved VCF Genotyper.
Significant run time improvements enable larger cohort analysis and reduced cost with comprehensive efficiency improvements of more than 2x the speed of the gVCF to msVCF workflow compared to DRAGEN 4.0. With improved msVCF configurability, filter out unnecessary variants while highlighting valuable insights with new outputs like inbreeding coefficient, Hardy-Weinberg equilibrium, and Excess Heterozygosity. Learn more about how gVCF Genotyper improvements are enabling genotyping variants at a population-scale in our recent article.
New DRAGEN Pipelines for Infectious Disease
Beta feature: DRAGEN Secondary Analysis pipelines for Urinary Pathogen ID/AMR Panel (UPIP) and Respiratory ID/AMR Panel (RPIP).
DRAGEN v4.2 enables bioinformaticians to explore Explify UPIP and RPIP pipelines on server with a command line interface. The UPIP/RPIP pipeline implements a precision metagenomics method using a k-mer classification tool. This is offered as DRAGEN's first metagenomics standalone tool.
UPIP panel and pipeline offers comprehensive detection and quantification of >170 pathogens, including fastidious, slow-growing, and anaerobic uropathogens and sexually transmitted microorganisms.
RPIP panel and pipeline offers comprehensive detection and quantification of >280 pathogens, including SARS-CoV-2, Influenza, RSV, and Mycobacterium and Legionella species.
For researchers interested in additional analysis and reporting functionality with an easy-to-use interface, check out the Explify UPIP and RPIP analysis applications on BaseSpace Sequence hub.
Additional Features
- ORA Compression Runtime improvements: Run ORA compression and DRAGEN map/align in parallel for a 15-30% speed increase from sequential processing.
- High Sensitivity Mode: 1) Ambiguous SNV calling in seg dup regions and 2) Enable low allele frequency (mosaic) calling with DRAGEN V4.2 high sensitivity mode (non-default).
- Sex Chromosome Mosaic Variant Support
- Structural Variance Improvements: Multigenome (graph) haplotypes guides alignments, revealing true breakpoint signatures to improve germline SV recall while maintaining high precision.
- New Systematic Noise Files + Alu regions Blocklists: significant FP reduction in somatic TO pipelines with updated systematic noise files and optional Alu regions blocklist.
- Fully Supported Amplicon Analysis: DRAGEN v4.2 fully supports amplicon analysis for SNV, RNA-fusion, CNV and SV.
- Enhanced Bulk-RNA QC metrics
- VCF Imputation: Enabled for haploid species, and chromosome X for diploid species.
Learn More
This blog highlights only five of the many key features in DRAGEN v4.2. Explore and learn more in the user guide available on our support page.
Visit the DRAGEN page to learn more about our technology .
Other Resources:
- https://precision.fda.gov/challenges/10
- https://precision.fda.gov/challenges/22/results
- Illumina Data on File, 2023
For Research Use Only. Not for use in diagnostic procedures