- Home
- News & Updates
- DRAGEN™ Enrichment App – Accurate, rapid analysis for germline and somatic exome experiments
-
DRAGEN
-
Product updates
- 09/09/2019
DRAGEN™ Enrichment App – Accurate, rapid analysis for germline and somatic exome experiments

Author: Eric Allen, Associate Director of Bioinformatics at Illumina
As part of the new DRAGEN v3.4 launch, the Illumina software development team has released a new BaseSpace-exclusive DRAGEN app – DRAGEN Enrichment v3.4. Combining the best of DRAGEN with Illumina’s legacy Enrichment 3 App, the DRAGEN Enrichment app provides ultra-rapid analysis and improved accuracy all at a lower cost per sample.
The DRAGEN Enrichment app is the preferable method for analyzing enrichment data with DRAGEN, delivering a full suite of enrichment specific metrics and reporting.
Here is what to know:
- The DRAGEN Enrichment App is faster and more accurate vs Enrichment (Isaac/Starling) and BWA Enrichment (BWA/GATK) apps, as demonstrated via the visuals below
- Variant Calling:
- Small variant calling - The app includes germline and somatic (low-frequency) small variant calling (tumor only); outputs VCF and gVCF in same analysis
- Note: Tumor-normal analysis can be conducted by first running the DRAGEN Enrichment app on all their normal and tumor samples, and then running the DRAGEN Somatic app on the resulting BAM files for the Tumor/Normal pairs.
- Copy number variant (CNV) calling – utilize CNV baseline files based on a panel of normals
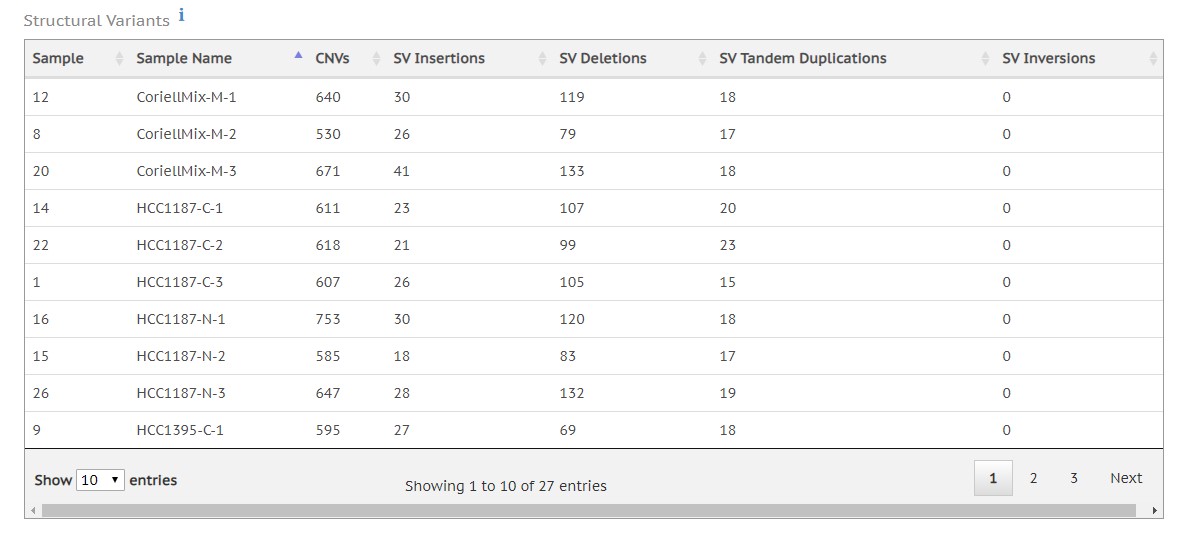
- Structural variant calling
- Small variant calling - The app includes germline and somatic (low-frequency) small variant calling (tumor only); outputs VCF and gVCF in same analysis
- Enrichment metrics generated:
- Read/base enrichment padded/unpadded
- Uniformity
- % bases covered at 1x, 10x, 20x, 50x
- Picard HsMetrics enabled by checkbox
- Variety of reference options supported, including hg19, GRCh38 and custom references
- Includes built-in targeted region BED files for common enrichment panels, and accepts custom targeted region BEDs
- Extensive reporting:
- In-browser, PDFs, and CSVs
- Single sample and aggregate reports
- Integrated variant annotation (Nirvana) and variant browser
The improved small variant calling over other available BaseSpace app solutions is shown below for one replicate of Coriell sample NA12878 with 106x depth:
| Analysis App | App Execution Time | DRAGEN-only Execution Time | SNV Recall | SNV Precision | Indel Recall | Indel Precision |
| DRAGEN Enrichment v3.4.5 | 16m 4s | 6m 50s | 95.04% | 99.49% | 86.90% | 92.18% |
| (Isaac/Starling) Enrichment v3.1.0 | 53m 20s | NA | 93.26% | 99.38% | 78.29% | 86.90% |
| BWA Enrichment v2.1.2 | 1h 23m 2s | NA | 90.66% | 99.78% | 72.85% | 89.44% |
• Example sample (s01-NFE-CEX-NA12878-demo.vcf) was prepared using Nextera Flex for Enrichment Library Preparation kit with dual indices and sequenced on a NovaSeq™ S2 flow cell: https://basespace.illumina.com/s/FaxWSm2X1gwO
• Variant accuracy comparison was performed using the Variant Calling Assessment Tool v3.2.0 app.
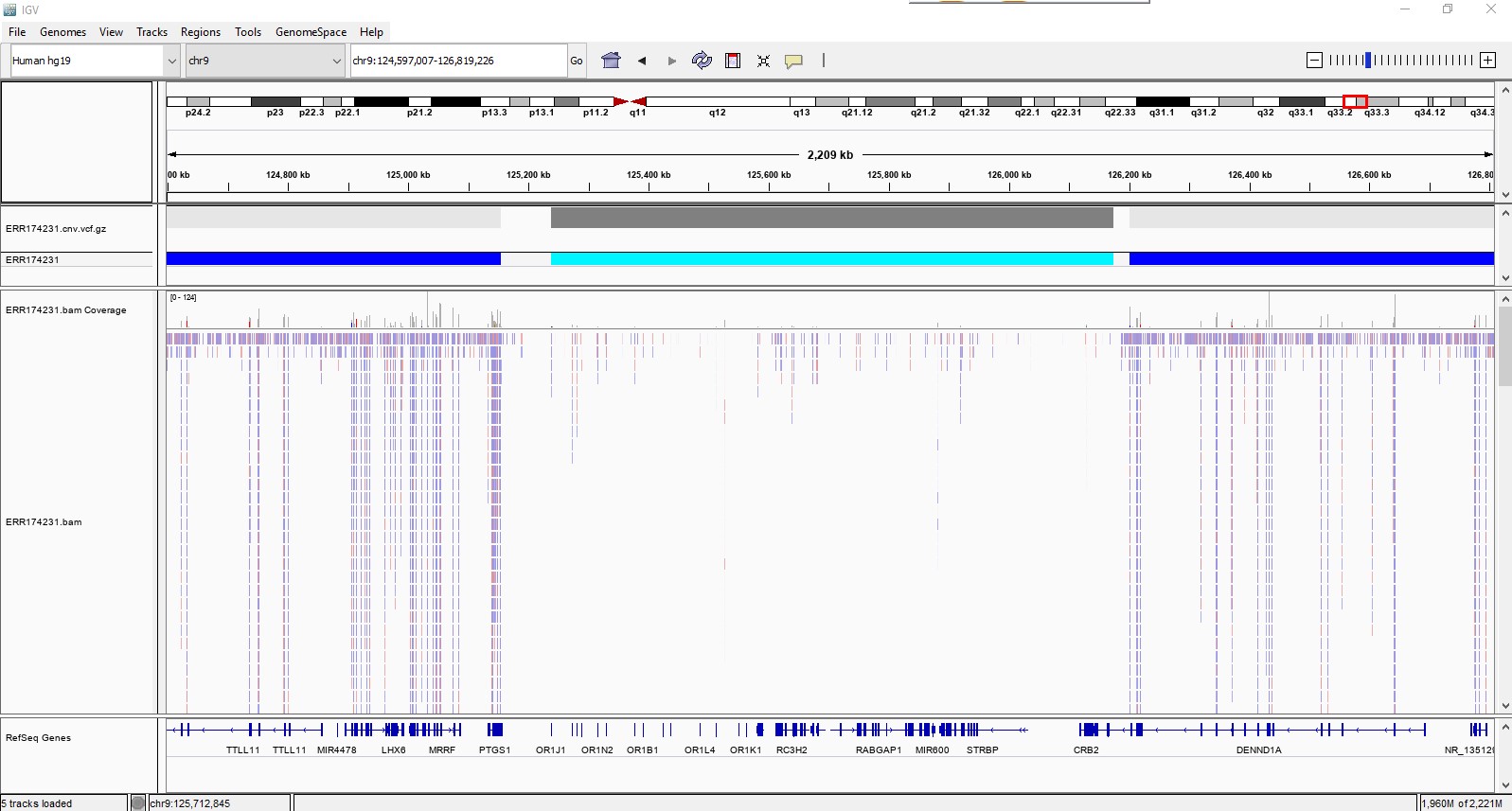
CNV calling is also enabled in the DRAGEN Enrichment app. The screenshot below from IGV shows a 937,697 bp CNV loss found in a melanoma cancer sample (Me01/ERR174231) around the chromosomal region chr9:125239269-126176965. The sample data was obtained from NCBI’s Sequence Read Archive (accession ERR174231) using the SRA Import BaseSpace App.
Somatic/low-frequency variant calling is also enabled. The table below demonstrates the usefulness of this somatic calling tool:
| Variant Type | Chr | Pos | Gene | Variant | HD753 - Expected VF (%) | HD753 – Measured VF (DRAGEN Enrichment) (%) |
| SNV Low GC | chr.3 | 178936091 | PIK3CA | E545K | 5.6 | 3.8 |
| SNV High GC | chr.19 | 3118942 | GNA11 | Q209L | 5.6 | 6 |
| Long Deletion | chr.7 | 55242464 | EGFR | ΔE746 - A750 | 5.3 | 3.3 |
| Long Insertion | chr.7 | 55248998 | EGFR | V769_D770insASV | 5.6 | 3.7 |
| SNV High GC | chr.14 | 105246551 | AKT1 | E17K | 5 | 5.7 |
Project: NovaSeq S4: Nextera Flex for Enrichment (HCC1187, HCC1395, HCC1954, HD753, Coriell Mixture). 1% VF cutoff
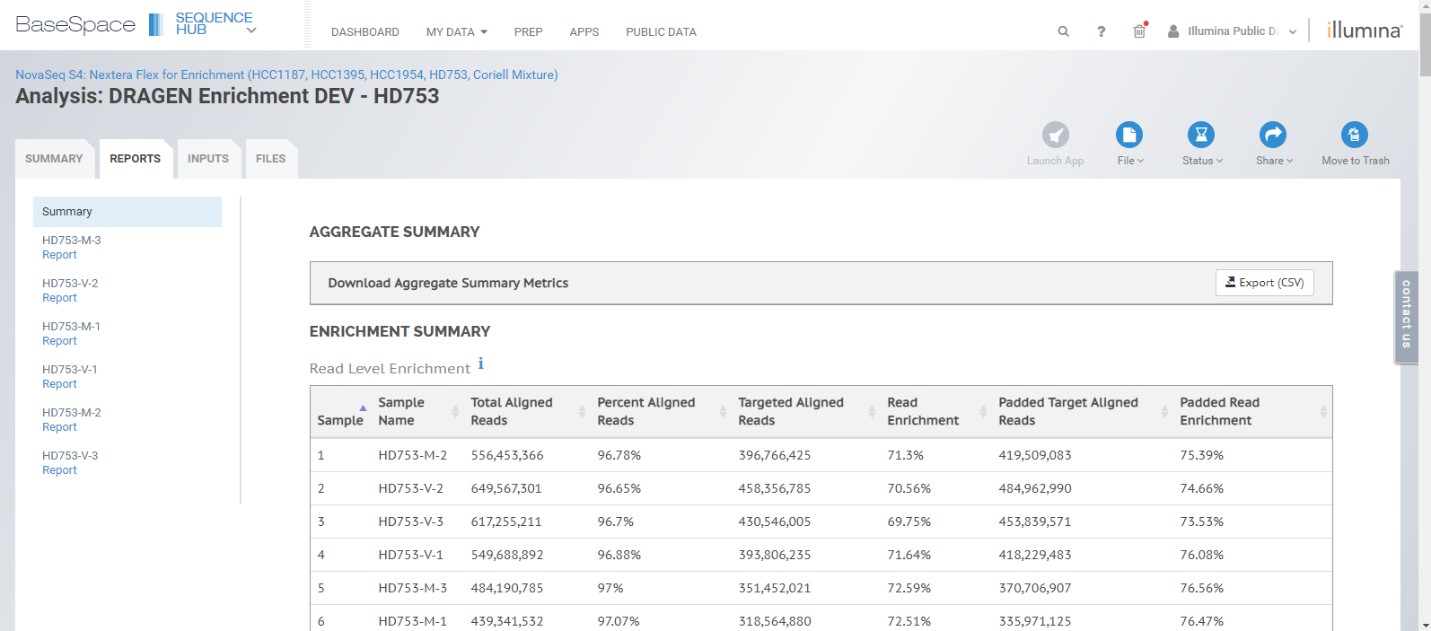
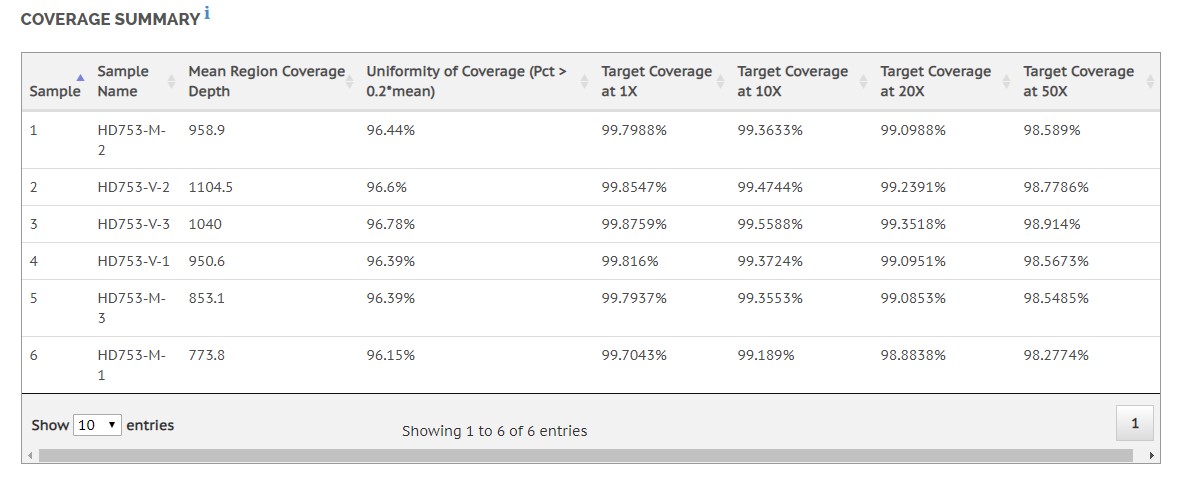
We’ve also incorporated many of the comprehensive metrics and reporting features built into the legacy Enrichment 3.1.0 app, including read-, base-, and target-level enrichment metrics, as well as the variant table for simple variant call browsing and filtering.
We hope this update enables you to discover new insights. Stay tuned for more app announcements, and let us know if you have any questions.