- Home
- News & Updates
- The Power of Precision: Benchmarking DRAGEN’s Somatic Variant Calling Accuracy
-
DRAGEN
-
Product updates
- 01/28/2026
The Power of Precision: Benchmarking DRAGEN’s Somatic Variant Calling Accuracy
Implementing next-generation sequencing at scale for oncology samples remains a persistent challenge for molecular pathology labs and those intending to implement in a clinical research setting. The inherent complexity of the tumor microenvironment, shaped by clonal heterogeneity and the unavoidable presence of "contaminating" normal cells, makes the detection of low-level somatic variants and tumor next-generation sequencing (T-NGS) an order of magnitude more challenging than identifying genetic disease markers.
As oncology advances push detection limits ever lower, accurately separating true low-frequency variants from background noise has become increasingly complex, requiring computational rigor beyond what most legacy open-source pipelines were built to deliver. To address this, the Illumina DRAGEN™ somatic pipeline was designed to deliver high sensitivity for low-VAF variants without compromising precision. By utilizing probabilistic noise models that characterize artifacts at runtime, a de Bruijn graph for accurate indel assembly, and a robust set of somatic-specific filters (including systematic noise files), DRAGEN effectively navigates 'noisy' genomic environments. But in a field where analytical rigor is paramount, internal benchmarks are only the beginning of the conversation. How does this performance translate to an independent, peer-reviewed publication when compared to other methods?
A recent publication in Genome Biology by Moon et al. (2025) of the National Cancer Center Korea provides some compelling answers, offering a deep dive into the performance of DRAGEN in a tumor-only pipeline scenario, using precisely titrated reference-standard DNA mixtures. By mixing homozygote hydatidiform mole DNA with heterozygote blood DNA, the researchers established a definitive 'truth set' to rigorously evaluate DRAGEN’s accuracy in detecting low-frequency somatic-like variants against a germline background.
The Analytical Challenge: Navigating the Noise
In a recent study published in Genome Biology by the National Cancer Center Korea, "Evaluation of false positive and false negative errors in targeted next-generation sequencing," Moon et al. (2025) set out to benchmark the accuracy of various NGS secondary analysis providers using precisely titrated reference-standard DNA mixtures. By admixing somatic variants at varying frequencies, they highlighted a stark reality: false positive (FP) error rates can vary by as much as 615-fold, and analytical sensitivity by 13.9-fold, depending on the provider and bioinformatics pipeline employed.
For the molecular pathologist, an FP is not just a data point; it’s a potential interpretation bottleneck that requires manual curation and literature review while a high Limit of Detection (LOD) risks missing actionable low-frequency mutations. In both scenarios, the lack of pipeline standardization can lead to inconsistent decision-making and compromised data integrity.
The Benchmark: DRAGEN Performance vs. Conventional Open-Source Stacks
Moon et al. conducted a head-to-head comparison between DRAGEN and the conventional pipelines (BWA-MEM + GATK Mutect2), as well as various other third-party solutions. Using reference-standard DNA mixtures to simulate the low-VAF (Variant Allele Frequency) environment of a real-world tumor, the results revealed a significant performance gap:
- Drastic Reduction in False Positives: In a direct comparison using Company CC’s data, DRAGEN reduced false-positive (FP) errors by 96.7%, dropping from 209.46 per Mb with the in-house pipeline to just 5.77 per Mb with DRAGEN (p = 0.0010). Furthermore, a conventional GATK-based workflow produced nearly four times as many FP errors as DRAGEN, demonstrating that DRAGEN’s integrated noise-suppression algorithms are significantly more adept at filtering out systematic artifacts.
- Superior Analytical Sensitivity: DRAGEN maintained high sensitivity for variant detection with a 95% detection threshold as low as ~1% to 1.6% VAF, a benchmark achieved on the Company CC (CancerSCAN™) and Company DD (TSO500) datasets at a median depth of 800x–1,000x. This significantly outperformed some in-house methods that had "blind spots" for low-frequency variants, with one provider failing to consistently detect variants below a 20% allele frequency.
- The Pipeline as a Performance Driver: The study confirmed that the secondary analysis engine, not just the sequencing chemistry, is a primary driver of accuracy. Even when using the same raw FASTQ data, DRAGEN and certain in-house solutions differed by up to 36.3-fold in FP rates, proving that algorithmic superiority is essential for reliable and reportable results.
- Resolving Complex Regions: One of the most significant findings involved the HLA (Human Leukocyte Antigen) genes. Mapping to these highly polymorphic regions is notoriously difficult, and the study found that many in-house providers suffered from persistent false negatives in these areas. In contrast, DRAGEN’s calls aligned closely with expected values, even in these complex regions, demonstrating the precision of its local assembly and alignment algorithms.
- Validation with Orthogonal Methods: The study utilized Single Base Extension (SBE) assays to resolve discrepancies between pipelines. When validating 22 discordant calls where DRAGEN and in-house pipelines disagreed, the SBE results confirmed that DRAGEN was correct in over half the cases by correctly filtering out the false positives that the in-house methods had misidentified.
The DRAGEN Impact: Standardizing Analytical Performance
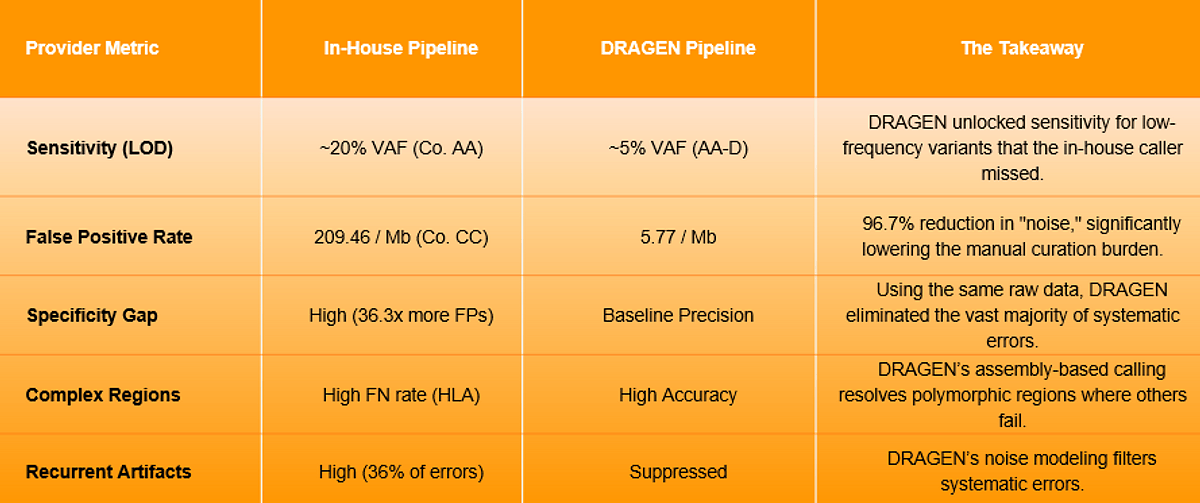
The study also identified 'recurrent FP-prone alleles': specific genomic sites highly susceptible to systematic errors. These sites accounted for over 36% of all errors in standard pipelines, creating a massive burden for clinical research teams. DRAGEN’s ability to model and suppress these artifacts at hardware-accelerated speeds allows labs to scale their throughput without a proportional increase in manual review time. By consolidating complex workflows into a single, highly optimized command line, DRAGEN simplifies pipeline management and scaling for clinical research applications, ensuring that high-volume testing remains both computationally fast and analytically precise.
Why the Algorithmic Gap Matters
The study demonstrates that precision and sensitivity are two sides of the same coin. The gap between a raw variant list and a meaningful report is defined by how a pipeline distinguishes true signal from background noise.
- Lowering the Noise Floor for Higher Sensitivity: Sensitivity, the ability to find a variant, is often limited by a pipeline's inability to manage noise. When pipelines use overly aggressive "hard filters" to avoid false positives, they inadvertently bury true low-VAF variants. DRAGEN’s probabilistic noise modeling and support for UMI collapsing, allows it to characterize artifacts at runtime, effectively lowering the noise floor. This allows the pipeline to "see" and confidently call low-frequency variants that other methods are forced to discard as noise.
- Eliminating Technical Debt and Manual Review: While sensitivity ensures you miss nothing, precision ensures you don't waste time on artifacts. A high false-positive rate creates "technical debt" for the molecular pathologist. DRAGEN’s noise modeling and specific set of filters prevent these recurrent artifacts from ever reaching the report, allowing labs to focus their expertise on interpretation rather than data cleanup.
- Stability Against Batch Effects: Custom in-house pipelines are often sensitive to "batch effects" where subtle variations in sequencing runs lead to inconsistent results. By adopting DRAGEN’s standardized architecture, laboratories ensure high reproducibility. This stability means that a sample processed today will yield the same high-quality results as a sample processed next month, providing a consistent analytical baseline for every patient.
- Navigating Genomic Complexity: Traditional linear aligners often struggle in "dark matter" regions like the HLA (Human Leukocyte Antigen), leading to false negatives. DRAGEN utilizes de Bruijn graph-based assembly to resolve these highly polymorphic regions, recovering true variants with high-fidelity accuracy where traditional methods see only ambiguity.
Final Thoughts
As molecular pathology labs move along the oncology continuum and begin to adopt panels larger in size and complexity, from small, targeted panels to Comprehensive Genomic Profiling (CGP) and Whole Genome Sequencing (WGS), the variant calling accuracy of bioinformatics pipelines becomes even more critical than ever. By leveraging DRAGEN and its integration with the broader Illumina informatics ecosystem, including Illumina Connected Insights, labs aren't just getting a variant list; they are getting a refined, analytically validated foundation for precision medicine research.
For those interested in diving into the technical specifics, we highly recommend reviewing the paper in Genome Biology and technical details on DRAGEN. For the recommended settings when running the DRAGEN somatic pipeline on tumor only panels, please refer to the DRAGEN recipe.
Ready to set the standard in your lab? Request a free trial of DRAGEN to get started.
References
Moon, et al. (2025). "Evaluation of false positive and false negative errors in targeted next generation sequencing." Genome Biology. DOI: 10.1186/s13059-025-03882-2
*Illumina internal data on file. 2025.
For Research Use Only. Not for use in diagnostic procedures.
M-GL-04039