- Home
- News & Updates
- DRAGEN v3.9 – Multi-cloud support, accuracy improvements, and more
-
DRAGEN
-
Product updates
- 08/06/2021
DRAGEN v3.9 – Multi-cloud support, accuracy improvements, and more
As we continue to unlock the power of the genome with new and advanced applications, the amount of data generated from next-generation sequencing (NGS) is rapidly expanding. It is estimated that by 2025, we would require 2-40 exabytes of storage for human genomic data1. Researchers and bioinformaticians need access to secure, high-performance computing and scalable storage to keep up with the vast amount of data. We aim to enable our customers to get faster genomic insights, so they focus on discovery instead of building and maintaining data processing and management infrastructure. Increasingly, we are seeing customers turning to the cloud to meet their needs for flexible scaling of genomic analysis and data management. To that end, we are excited to announce that DRAGEN v3.9 brings Microsoft Azure support, making DRAGEN even more accessible on the cloud. Of course, it wouldn’t be a DRAGEN release without a whole slew of additional new features and enhancements across the portfolio.
>>For a deeper dive, sign up for our upcoming webinar and check out the software release note here.
Let's look at some of the new and exciting improvements in DRAGEN v3.9.
DRAGEN availability on Microsoft Azure
Microsoft recently released their NP series of FPGAs in four regions globally – East US 2, West US, West Europe, and Southeast Asia. And now with the DRAGEN v3.9 release, you can run DRAGEN on Microsoft Azure and see whole genome run times of ~40 mins at 30x coverage. 2 Check out the Marketplace listing here. You’ll need a DRAGEN license from your Illumina sales rep to get started. Reach out to insidesales@illumina.com to learn more.
RNA Variant calling
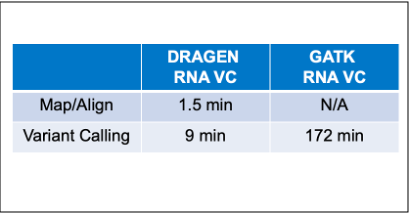
The legendary DRAGEN variant calling accuracy is now available in our RNA pipeline. This new feature leverages the DRAGEN RNA Mapper and Somatic Variant Caller to directly call variants from RNA samples. DRAGEN RNA VC is 19 times faster than GATK while providing comparable accuracy.3
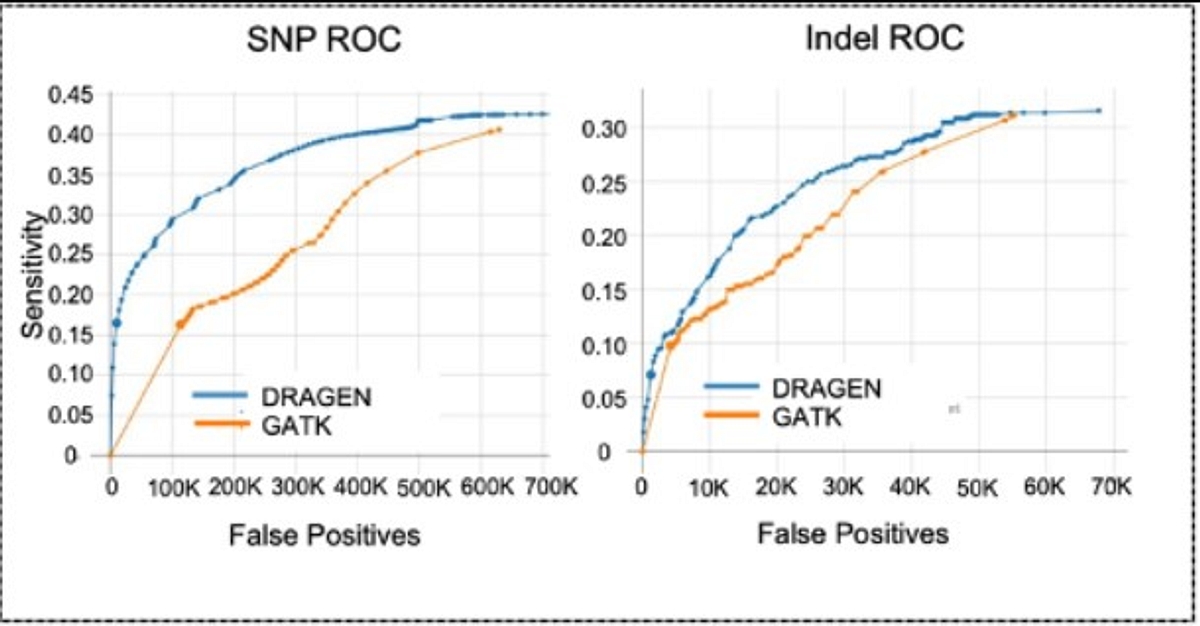
Figure 1- RNA VC ROCs showing sensitivity vs false positive count on a HCC1187 WTS sample. Germline calls from matched DNA sample used as truth set. Both DRAGEN and GATK use RNA-mapped DRAGEN BAMs as input.4
New Biomarker - Homologous Recombination Deficiency (HRD)
HRD is an emerging biomarker for Poly (ADP-ribose) polymerase (PARP) inhibitors in multiple cancer types.5 The new DRAGEN HRD module computes the HRD score based on Sztupinszki methodology6 from whole genome tumor-normal and tumor-only somatic CNV calling. The caller has high concordance with the widely-used microarray approach.7
HRD is added to the expanding list of oncology biomarkers DRAGEN now supports including tumor mutational burden (TMB), Microsatellite Instability (MSI), and HLA typing.
Accuracy gains
With DRAGEN v3.9, we have made accuracy gains across the whole portfolio.
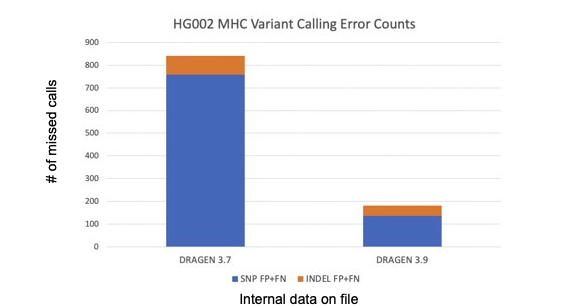
- DRAGEN 3.7 introduced graph-based read mapping. v3.9 further improves read mapping and SNV/indel variant calling (VC) accuracy in the Major Histocompatibility Complex (MHC) with up to 80% accuracy increase.
- V3.9 includes updates to alternative (ALT) contig handling and graph reference updates, which leads to improved mapping and variant calling accuracy for hg19/hg38 genomes.
- With the v3.9 release, we are also providing access to an alpha version of a powerful machine learning tool that refines small variant calls, leading to a ~33% Germline SNV accuracy improvement
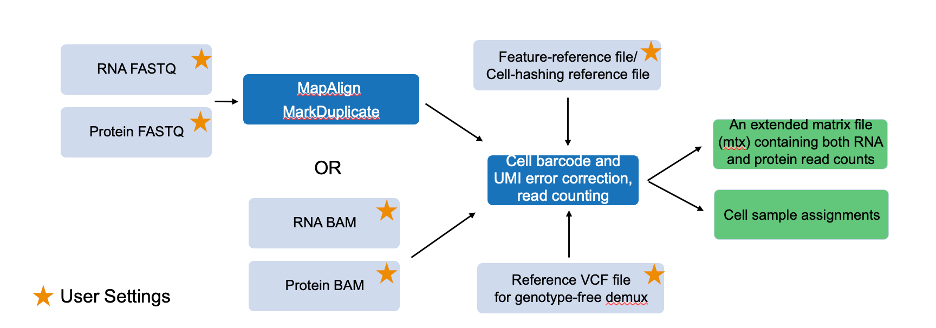
Single-Cell Updates
The new single-cell RNA (scRNA) pipeline features include:
• Protein/Antibody measurements: Measure the expression of cell-surface proteins or antibodies along with the expression of genes from the same cell.
• Cell Hashing: A new technique for assigning sample identities to cells in a single-cell RNA experiment. It is based on tagging a part of the reads from each cell with a specific oligo sequence.
Beyond these key updates, we’ve also streamlined our CNV caller, enabled gene fusion detection in repetitive regions, added features to DRAGEN ORA compression, and much more!
To learn more about DRAGEN v3.9, register for our upcoming webinar here.
If you are interested in getting started with DRAGEN on Azure, contact your Illumina team.
For Research Use Only. Not for use in diagnostic procedures.
- https://sapac.illumina.com/company/news-center/blog/solving-for-the-information-gap-in-genomics-breakthroughs.html
- https://science-docs.illumina.com/documents/Informatics/dragen-v3-accuracy-appnote-html-970-2019-006/Content/Source/Informatics/Dragen/dragen-v3-accuracy-appnote-970-2019-006/dragen-v3-accuracy-appnote-970-2019-006.html
- Data on file, Illumina, Inc., 2021
- Data on file, Illumina, Inc., 2021
- https://www.nature.com/articles/s41598-020-59671-3
- https://www.nature.com/articles/s41523-018-0066-6
- Microarray HRD Scores https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4443545/