- Home
- News & Updates
- DRAGEN™ v4.0: Powering Single-Cell Genomics Insights
-
DRAGEN
-
Product updates
- 09/21/2022
DRAGEN™ v4.0: Powering Single-Cell Genomics Insights
Single-cell sequencing is a next-generation sequencing (NGS) method that investigates gene expression of individual cells and showcases cell-to-cell variation. Gene expression profiling allows us to study cell function, both in a normal and pathologic state, and processes such as differentiation, proliferation, and tumorigenesis. Single-cell sequencing powers insights into the distinct biology and responses of individual cells and cellular populations in complex tissues.
Advances in single-cell genomics have led to a need to increase experimental scale, resulting in challenges related to data processing, data management, and data storage. DRAGEN provides accurate, comprehensive, efficient processing of NGS data, and can fully process a 34x whole human genome in ~30 minutes. To handle the storage demands of large NGS data files, DRAGEN Original Read Archive (ORA) technology provides lossless 5x compression of FASTQ files in ~8 minutes for 50 GB to 70 GB FASTQ files. DRAGEN v3.7 introduced an integrated single-cell RNA-seq pipeline that goes from base calls to cell-by-gene-expression matrix with a single user touchpoint.
In addition to our existing single-cell RNA-seq pipeline, DRAGEN v4.0 now introduces two new, innovative pipelines for discovery research customers:
- Single-cell ATAC-seq
- Single-cell multiomics: (RNA-seq + ATAC-seq)
The DRAGEN single-cell ATAC-seq pipeline enables single-cell resolution of chromatin accessibility for better insight into cell type, gene expression, and regulatory mechanisms. The DRAGEN single-cell multiomics pipeline allows the processing of combined single-cell ATAC and single-cell RNA datasets generated from the same set of biological cells, enabling simultaneous profiling of transcriptome and epigenome. The DRAGEN single-cell multiomics pipeline can process data sets from single-cell RNA-Seq and ATAC-Seq reads to a cell-by-feature count matrix. The workflow starts with FASTQ files as inputs and produces a joint count matrix in sparse format that for each cell barcode contains information about how many UMIs correspond to each gene and how many ATAC fragments intersect each peak. The runtime is ~12 minutes for 50M RNA reads, 200M ATAC reads, 2500 cells.
DRAGEN single-cell multiomics pipeline:
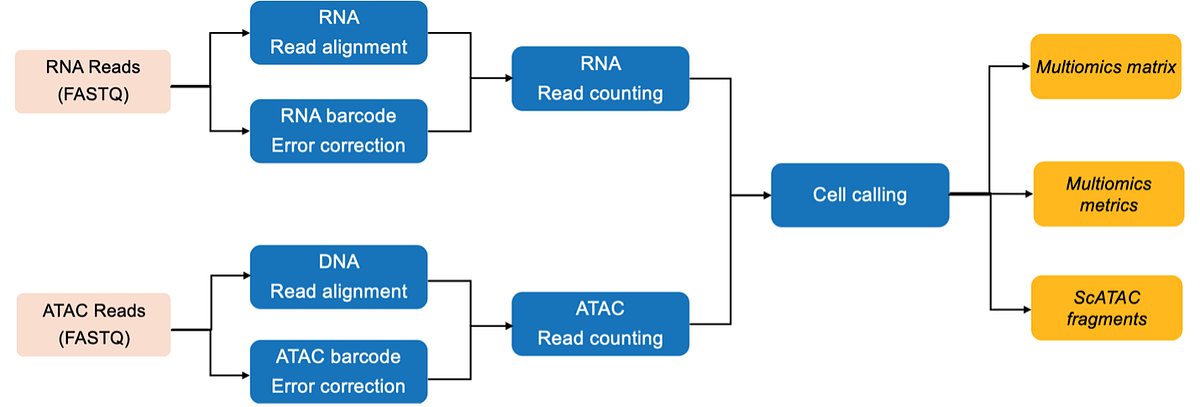
The DRAGEN single-cell ATAC-seq pipeline is available on-premise, on-instrument (NextSeq 1000/2000), or cloud solutions (BaseSpace™ Sequence Hub). The DRAGEN single-cell multiomics pipeline is available on-premise.
DRAGEN single-cell multiomics pipeline can be enabled using:
--enable-rna=true
-–enable-single-cell-rna=true
–-enable-single-cell-atac=true
Additionally, only single-cell ATAC pipeline can be enabled using:
--enable-single-cell-atac=true
Learn More:
The DRAGEN 4.0 release is packed full of comprehensiveness, accuracy, and efficiency gains, and this blog has only touched the surface.
For an in-depth summary of all DRAGEN 4.0 features, view our user guide.
Visit the DRAGEN page to learn more about our technology and other updates, including our latest v4 server release.
Illumina Connected Analytics Single-Cell Resources:
Illumina Connected Analytics - Interactive Analysis for a Single Cell RNA Workflow
Other Resources:
For Research Use Only. Not for use in diagnostic procedures.