- Home
- News & Updates
- HIVE single cell RNAseq libraries data from Honeycomb Biotechnologies are now available on Illumina BaseSpace™ Sequence Hub demo data page!
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 01/06/2023
HIVE single cell RNAseq libraries data from Honeycomb Biotechnologies are now available on Illumina BaseSpace™ Sequence Hub demo data page!
Biological resolution at the level of individual cells is powering the next phase of precision health. The HIVE scRNAseq Solution from Honeycomb Biotechnologies integrates sample storage and single cell profiling into a complete workflow. In this blog, we are presenting NextSeqTM 2000 sequencing run data for HIVE scRNAseq libraries.
The HIVE scRNAseq Solution is a portable, handheld, single-use device that enables gentle capture, RNA stabilization without fixation, and easy processing for the analysis of single-cell samples without specialized instrumentation. The design of the HIVE is ideal for the recovery of fragile cells, loading of large sample volumes, and efficient processing of numerous samples in parallel. In this blog, we are presenting NextSeqTM 2000 sequencing run data for HIVE scRNAseq libraries. The HIVE scRNAseq libraries are also compatible with other Illumina platforms like the NextSeq 500/550 and the NovaSeqTM 6000 (https://honeycombbio.zendesk.com/hc/en-us/articles/4738507106587-Recommended-Sequencers-).
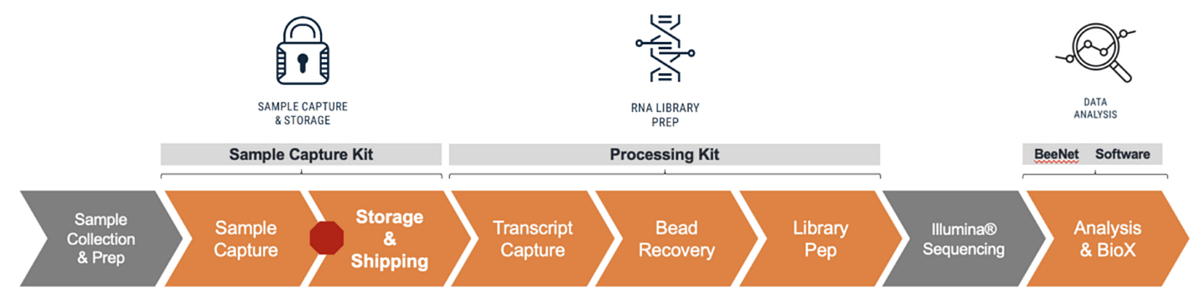
The HIVE scRNAseq workflow
The HIVE uses an array of 65,000 picowells that are preloaded with uniquely barcoded 3’ transcript capture beads to quickly isolate single cells. Once loaded, the Cell Preservation Solution is added to stabilize RNA and lock in molecular signals. Cell-loaded HIVEs can be shipped or stored until ready for Honeycomb’s simplified and scalable HIVE processing and library preparation workflow (figure 1). Quality control metrics (genes, transcripts, % mitochondrial reads), marker gene expression, and cell-type proportions are not significantly different between freshly processed cells and cells stored in the HIVE device for up to 9 months at -20°C.
HIVE scRNAseq run available on BaseSpace™ Sequence Hub
To view these HIVE scRNAseq libraries run on NextSeq 2000, please access the BaseSpace Sequence Hub and import the sequencing run using the link provided below. These demo data are free to use on the BaseSpace platform (see our previous post - https://developer.illumina.com/news-updates/demo-data-on-illumina-basespace-sequence-hub) and can be used for direct comparison with your own single cell sequencing data generated with HIVE scRNAseq.
To understand how to evaluate your sequencing run, see our post: https://developer.illumina.com/news-updates/basespace-sequence-hub-does-my-sequencing-run-look-good
Link to run: https://basespace.illumina.com/s/gEAzvbzEA8y4
Link to project: https://basespace.illumina.com/s/Yfr34h5YKo0w
HIVE scRNAseq libraries were sequenced on the NextSeq 2000, with a read length of 72 + 8 + 8 + 50. Figure 2 showcases the sequencing data by cycle showing Q30 and percentage base for the HIVE scRNAseq libraries on the NextSeq 2000 run. HIVE libraries require custom sequencing primers, which are included in the HIVE Processing kit, and were added to the reagents cartridge as explained in the NextSeq 1000/2000 documentation. 5% PhiX (v/v) can be spiked in the pool of libraries to help with the base diversity. Honeycomb recommends that spiking in PhiX is optional. If PhiX is spiked in, the run requires the addition of standard Illumina sequencing primers in addition to the HIVE customer primers.
Here, we demonstrate the combined workflow of HIVE scRNAseq and Illumina sequencing, working with cells isolated from a renal cell carcinoma (RCC) sample. A solid tumor biopsy from an adult male, pre-treatment, was dissociated by a mechanical and enzymatic method. Freshly dissociated cells were either loaded directly into the HIVE and stored for 20 weeks at -20C (figure 3, sample 4 to 6) or cryopreserved and stored in liquid nitrogen, also for 20 weeks, prior to loading into the HIVE (figure 3, samples 1 to 3).
Stay tuned for another post with a deep dive on this data set demonstrating the stability of storage with the HIVE versus the progressive degradation of sample quality with cryopreservation, including the progressive loss of immune cells.
Honeycomb & Illumina: Expanding Access to Single-Cell RNAseq
Stable HIVE storage enables remote sample collection sites with centralized processing, and multi-site collaborations, longitudinal studies, time-courses, and sporadic or end-of-day samples. Additionally, the robust recovery of high-quality biological data for fragile cell-types, such as neutrophils, neurons, hepatocytes, and epithelial cells, is possible with the HIVE compared to workflows with cryopreservation or microfluidics droplet-based technologies where these populations can be destroyed. The HIVE propels forward clinical research using blood, bone marrow, fine needle aspirates, and other minimally invasive biopsies in the context of infectious diseases, blood cancers, allergy/asthma, and auto immune diseases. Honeycomb Biotechnologies together with Illumina aims to expand single-cell opportunities across basic, translational, and clinical research throughout the world.
Additional resources:
Honeycomb links: Primary data analysis of demultiplexed FASTQ files with BeeNet after Illumina sequencing.
Example HIVE scRNAseq data set for review and download.
NextSeq 2000: https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000.html
To answer your questions regarding preparation, sequencing and analysis of HIVE libraries, contact Honeycomb Biotechnologies support (support@honeycomb.bio). For questions about your sequencing run, contact Illumina (techsupport@illumina.com).
For Research Use Only. Not for use in diagnostic procedures.