- Home
- News & Updates
- MobiDrop MobiCube Single Cell Multiomics Libraries data on the NovaSeq™ 6000 now available on BaseSpace™ Sequence Hub (BSSH)
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 05/24/2023
MobiDrop MobiCube Single Cell Multiomics Libraries data on the NovaSeq™ 6000 now available on BaseSpace™ Sequence Hub (BSSH)
Today we are delighted to showcase new data with the MobiDrop single-cell libraries on BaseSpace demo data page.
Single-cell sequencing technologies can be used to detect the transcriptome and other multi-omics such as the full-length V(D)J sequences of single lymphocytes. They can show the functional differences and evolutionary relationships of various cells. MobiDrop is a single-cell company who develops revolutionary gel beads and an advanced microfluidic system that enables the barcoding of tens of thousands cells within minutes, providing flexible, high-throughput, and easy-to-use solutions for life sciences, drug development and precision medicine.
MobiDrop single-cell workflow with Illumina sequencing
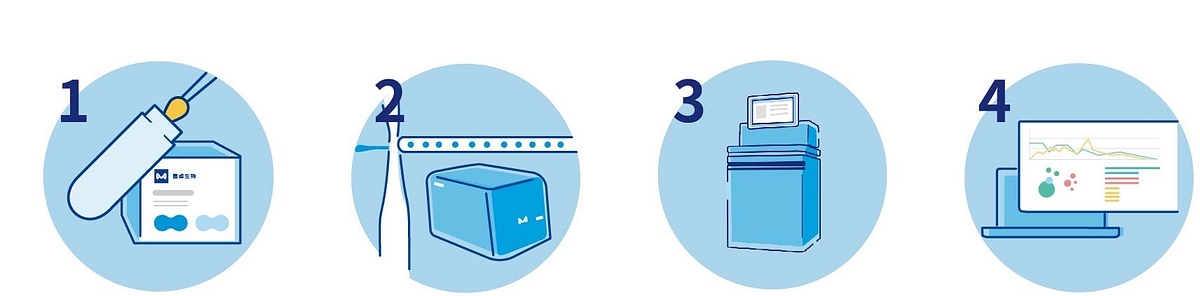
Figure 1: MobiDrop single-cell library workflow paired with Illumina sequencing. These libraries are compatible with all llumina® platforms, but we recommend using NextSeq™ 550/1000/2000 and NovaSeq™ 6000/X to be able to sequence multiple samples per run.
Sequencing set up recommendations
Table 1: Example sample throughput for MobiCube single-cell sequencing assays on Illumina sequencing systems
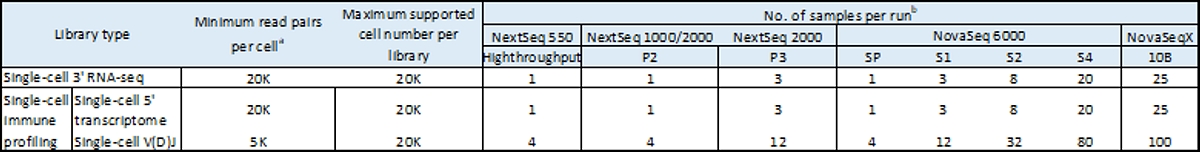
a. Minimum read recommendations from MobiDrop. 20K read pairs per cell or 40K individual reads, 20K from Read 1 and 20K from Read 2. Adjust sequencing depth according to desired performance or application. Up to 96 unique dual indexes are supported. The sequencing saturation metric and curve in the MobiVision reports can be used to optimize sequencing depth for specific sample types.
b. Number of samples per run recommendations from MobiDrop. Actual performance parameters may vary depending on the sample type, the sample quality, and the filtered clusters. See the specification sheet for the respective sequencing system for details.
Table 2: Recommended read configuration for MobiCube single-cell libraries
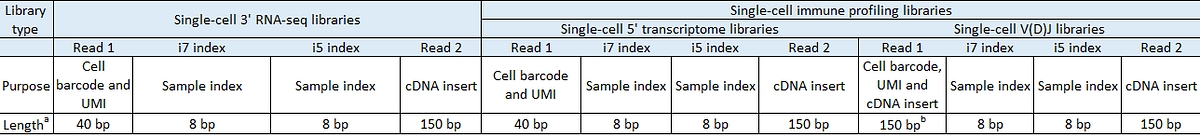
a. Shorter transcript reads may result in reduced transcriptome alignment rates. Cell barcode, unique molecular identifier (UMI), and sample index reads should not be shorter than recommended. MobiDrop has validated up to 2*150bp run for these libraries.
b. Sequencing length can be adjusted according to the sequencing kit utilized, but should not go below 40 bp. While it is possible to process a sequencing length between 40 bp and 150 bp in MobiVision, this may lead to a reduced number of identified V(D)J sequences.
MobiDrop single-cell run available on BSSH demo data page
To view a set of sequencing runs for MobiDrop libraries run on NovaSeq 6000 S4 flow cells, please visit BaseSpace™ Sequence Hub. MobiCube single-cell 3’ RNA-seq libraries (https://en.mobidrop.com/single-cell-multiomics) were prepared from peripheral blood mononuclear cells (PBMCs) and mouse tissues, and MobiCube single-cell V(D)J libraries (including paired 5’ RAN-seq and V(D)J libraries) were prepared from PBMCs, according to the MobiCube® High-Throughput Single-Cell 3’RNA-Seq Kit v2.0 User Guide and MobiCube High-Throughput Single-Cell V(D)J Kit User Guide, respectively. Libraries were sequenced on a NovaSeq6000 targeting at least 20K reads per cell and 10% PhiX was spiked in.
Check our previous blog post on our Demo Data section in BaseSpace Sequence Hub for additional details on how to access the run data on the demo data page.
Below is the link to directly import the runs and project folders into your account. These runs can be found under the category “Multiomics” and “single cell”. Because these are public data sets, these data sets are free and do not count against storage limits.
You can use the demo data run to compare with your own MobiDrop sequencing run. See our first blog post in this series on evaluating sequencing runs for additional details on how to estimate sequencing your run quality.
Run link: https://basespace.illumina.com/s/tOuyrWPda69M
Project link: https://basespace.illumina.com/s/Jd6XcMZ1d6Zx
Access MobiVision software html report
html reports generated with the MobiVision software (MobiDrop, Inc.) are available under the BaseSpace project. Select the Report in the “Analyses” category of the Project, then select “Files” where the html Web Summaries are available. They need to be downloaded for visualization.
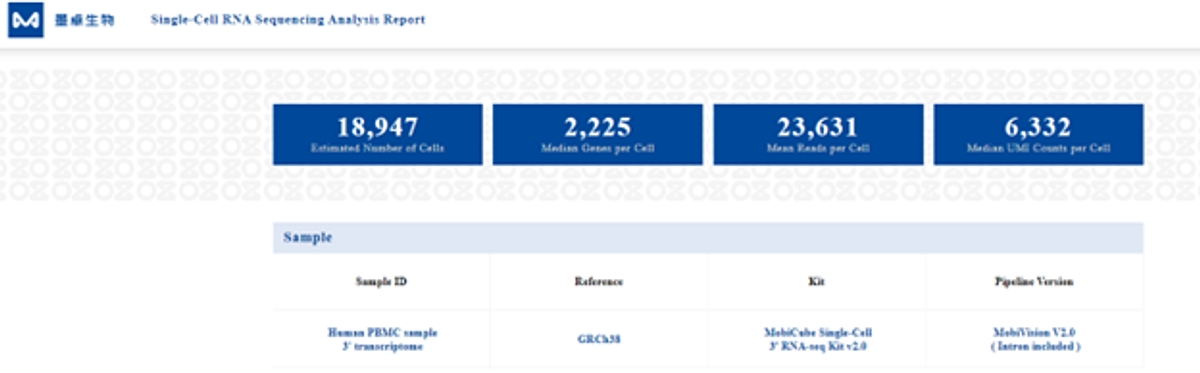
Figure 2: output from the MobiVision software
Additional resources
If you need further assistance in sequencing MobiDrop single-cell libraries, you can contact the Illumina® (techsupport@illumina.com) and MobiDrop (contact@mobidrop.com) support team.
Other posts from the blog series: new run data publicly available on BaseSpace™ Sequence Hub (BSSH).
Special thanks to Xianghua Hu from Illumina GC marketing team for enabling this partnership with MobiDrop.
For Research Use Only. Not for use in diagnostic procedures. M-GL-01825