- Home
- News & Updates
- New England Biolabs NEBNext Immune Sequencing Kit data now available on the new NextSeq™ 2000 600 cycles kit
-
BaseSpace™ Sequence Hub
-
Product updates
-
News
- 01/17/2023
New England Biolabs NEBNext Immune Sequencing Kit data now available on the new NextSeq™ 2000 600 cycles kit
Researchers are efficiently exploring immune responses with a new bulk RNA sequencing workflow that generates full-length reads of somatically recombined B and/or T cell receptor V(D)J genes. The discoveries incorporating this unique view are shedding light on previously unanswerable questions surrounding immune response to infection and therapeutic intervention.
Immune repertoire is a term used to define the sum of an individual’s expressed T and B cell receptors (TCRs and BCRs, respectively). It is a dynamic state, encompassing the scope of an immune response. The TCR/BCR expression profile contains a wealth of information about the mysteries of immune responses in health and disease from both recent and past exposures. The information can be used to tease out immune cell involvement in autoimmune disease or to evaluate a host immune response to vaccination. Also, because different diseases (and even disease severity) can lead to a specific immune profile, it is clinically relevant due to potential utility in directing treatment choices and predicting treatment efficacy.
With the NEBNext Immune Sequencing Kit (Human) and NEBNext Immune Sequencing Kit (Mouse), scientists at New England Biolabs have taken immune repertoire sequencing to a new level with a workflow that enables bulk mRNA sequencing of the variable regions of BCR heavy and light chains and TCR α and β chains in humans or mice generating full-length UMI-tagged sequences.
In this blogpost, we are showcasing NextSeq 2000 data with the NEBNext Immune Sequencing Kits, using the newly launched 600 cycles sequencing kits
Library preparation
To better understand the effects of input RNA quantity on required sequencing depth and clonotype saturation, 32x NEBNext Immune Sequencing libraries were prepared with Human PBMC total RNA and Mouse spleen RNA, ranging from 10 ng to 1000 ng. The NEB libraries were prepared such that BCR heavy chain (HC) and light chain (LC), as well as TCR alpha (α) and beta (β), were multiplexed together and separately. Replicate libraries were prepared under each condition to evaluate the technical reproducibility.
Accessing sequencing run
NEBNext Immune Sequencing Kit were sequenced on the NextSeq 1000/2000 P2 300M Reagents (600 Cycles) catalog number 20075295 and on NextSeq 1000/2000 P1 Reagents (600 Cycles) catalog number 20075294. To view this run on NextSeq2000 600 cycles kit, please access the BaseSpaceTM Sequence Hub and import the sequencing run using the link provided below.
Run link: https://basespace.illumina.com/s/AdTEXiU9iEIK
These demo data are free to use on the BaseSpace Sequence Hub platform (see our previous post - https://developer.illumina.com/news-updates/demo-data-on-illumina-basespace-sequence-hub)
Evaluation of the sequencing run
To understand how to evaluate your sequencing run, see our previous blogpost "Does my sequencing run look good?"
NEBNext Immune Sequencing Kit libraries were sequenced with a read length of 301 + 8 + 8 +301 on the NextSeq1000/2000 P1 100M Reagents (600 Cycles), catalog number 20075294. The NEB library pool was loaded at 650 pM on the NextSeq2000 run and the run is meeting Illumina specifications (https://www.illumina.com/syste...).
Due to the lower base diversity (figure 1) present in specific regions of UMI and full-length antibodies, 10% PhiX (v/v) was spiked in the pool of libraries to increase base diversity.
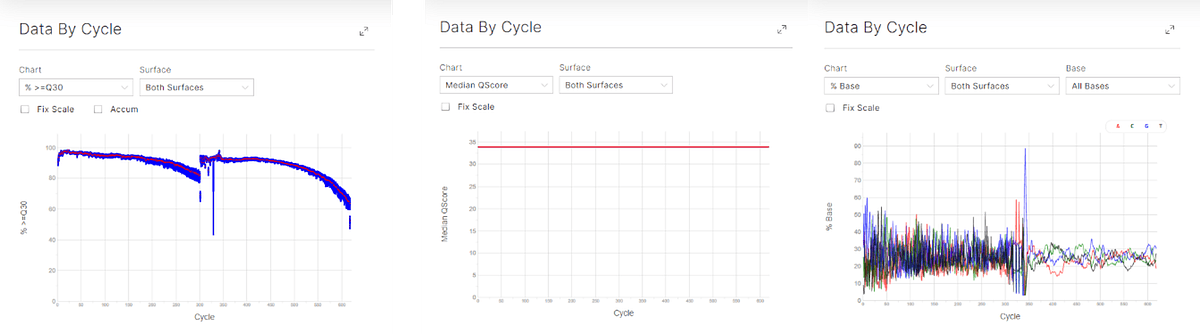
Figure 1: sequencing data by cycle showing the data by cycle for Q30 (left), median Q-score by cycle (middle) and percentage base by cycle (right).
Analysis
Project link contains the analyzed data: https://basespace.illumina.com/s/yI3vmxkEJ9ua
The results showed premium quality in sequencing metrics, including high quality in sequencing reads, vast majority of reads successfully assembled to full length V(D)J sequences and high confidence in clonotype annotation.
Learn more:
https://www.neb.com/products/e6320-nebnext-immune-sequencing-kit-human#Product%20Information
https://www.neb.com/products/e6330-nebnext-immune-sequencing-kit-mouse#Product%20Information
https://www.illumina.com/syste...
https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000/specifications.html
https://www.illumina.com/areas-of-interest/immunogenomics.html
Support contact information:
To answer your questions regarding preparation and sequencing of NEB immune repertoire libraries, contact NEB’s dedicated Technical Support team online (https://www.neb.com/forms/neb-...), via email (info@neb.com), or by phone (1-800-632-7799).
For questions about your sequencing run, contact Illumina (techsupport@illumina.com).
For Research Use Only. Not for use in diagnostic procedures.
Special thanks to our colleagues at NEB for providing library prep reagents and controls for this experiment and analyzing the IR-data. Hat tip to Illumina Scientists Robin Bombardi and Qiyuan Han and the Emerging Apps team for sequencing these runs for this collaboration.