- Home
- News & Updates
- Post-sequencing auto-launch DRAGEN™ single cell RNA analysis now enabled for Singleron GEXSCOPE single cell library on Illumina NextSeq™ 2000.
-
DRAGEN
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 09/19/2023
Post-sequencing auto-launch DRAGEN™ single cell RNA analysis now enabled for Singleron GEXSCOPE single cell library on Illumina NextSeq™ 2000.
In collaboration with Singleron Biotechnologies, here we show a streamlined workflow with auto launching onboard DRAGEN Single Cell RNA analysis after sequencing a Singleron GEXSCOPE single cell library on an Illumina NextSeq 2000 instrument.
Single cell RNA sequencing (scRNA-seq) enables profiling the transcriptome at single cell resolution. In comparison to traditional bulk RNA sequencing, the massive dataset and complexity of the assay bring additional challenges in the downstream data QC and secondary analysis. These become even bigger challenges when labs favor on-site analysis and don’t have high performance computing resources. Illumina NextSeq 2000 sequencer is equipped with FPGA chips that can provide robust and speedy onboard DRAGEN single cell analysis solutions for such scenarios.
Singleron Biotechnologies is an Illumina partner who develops single-cell multi-omics products for both research and clinical use. Singleron GEXSCOPE Single Cell RNA Library Kit is a complete solution for partitioning tissues into single cells and constructing next generation sequencing (NGS) libraries for single cell mRNAs. Here we show a streamlined workflow with auto launching onboard DRAGEN Single Cell RNA analysis after sequencing a Singleron GEXSCOPE single cell library on an Illumina NextSeq 2000 instrument. This workflow enables fast and accurate single cell analysis following a sequencing run completion with no intermediate manual touch points (Figure 1).
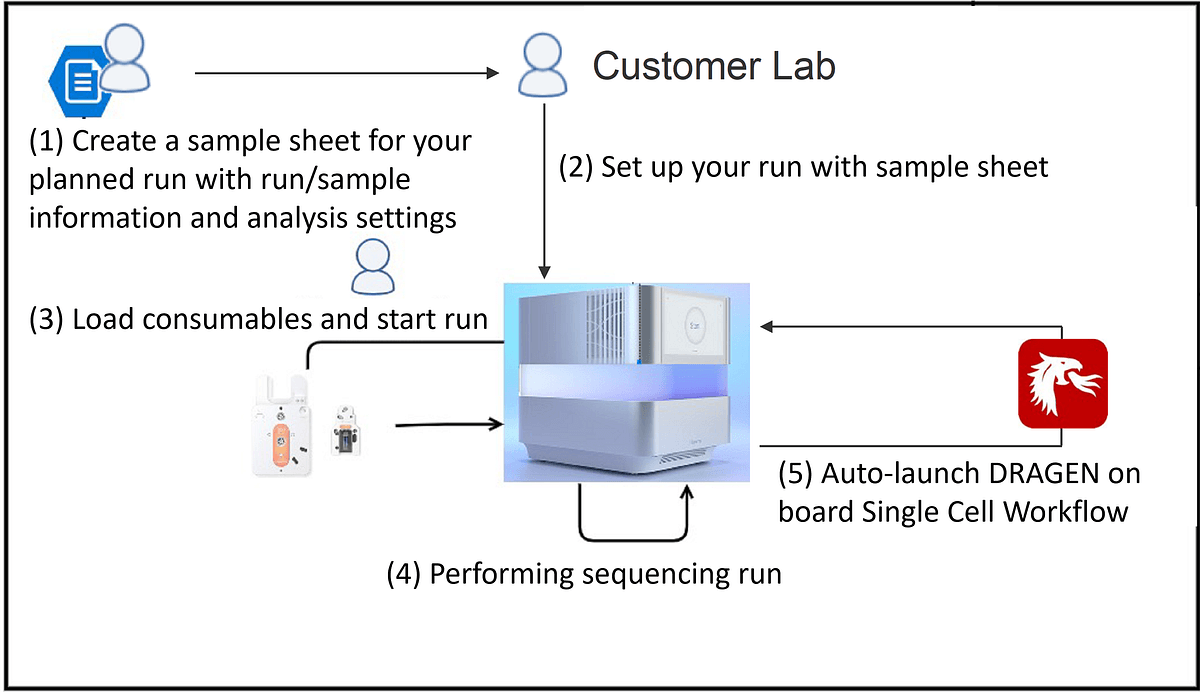
Figure 1. A streamlined workflow from sequencing Singleron GEXSCOPE single cell library on NextSeq 2000 to auto launching onboard DRAGEN Single Cell RNA analysis.
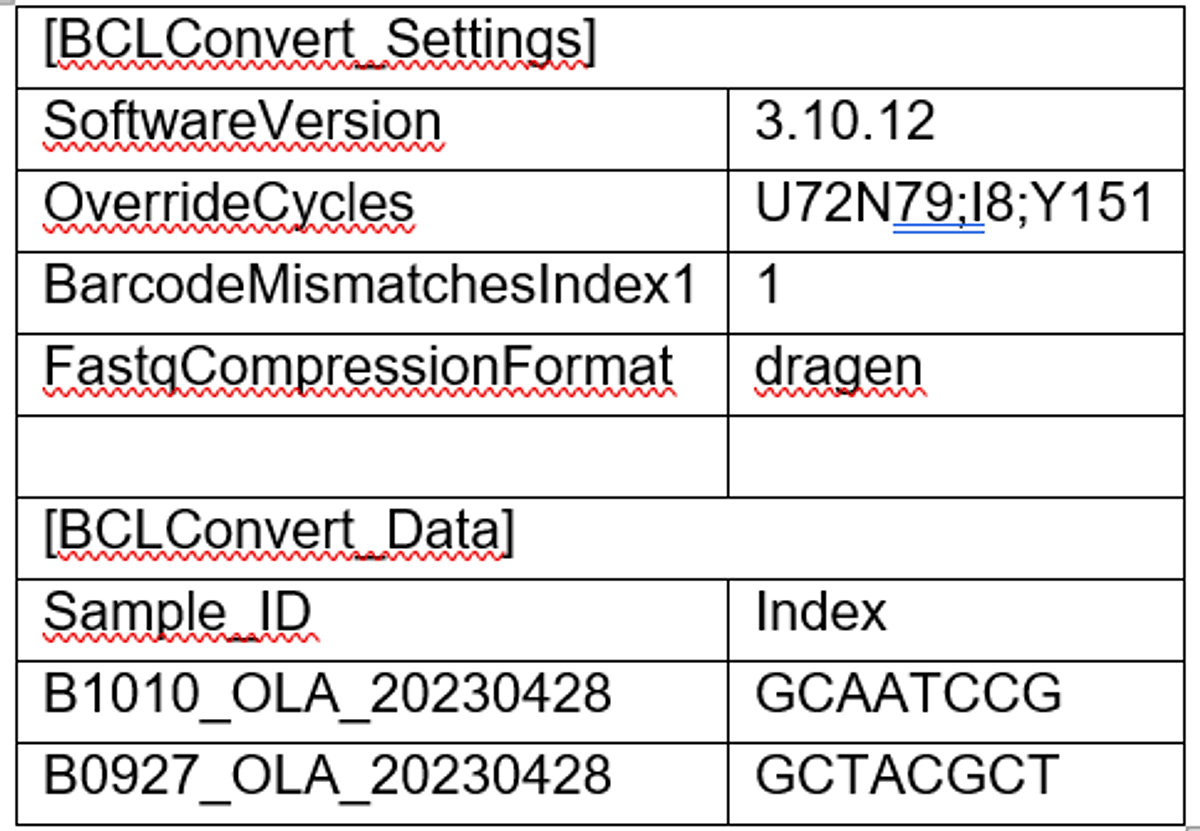
To present an example study, we conducted a scRNA-seq study to show the ease-of-use, speed, and robustness of this workflow. The sequencing library is a pool of two Singleron GEXSCOPE Single Cell RNA libraries. When executing this workflow, researchers start by generating a v2 Sample Sheet. The Sample Sheet has 3 major components including: run information and sequencing mode setting (Table 1), BCL Convert settings (Table 2), and DRAGEN Single Cell RNA pipeline settings (Table 3). The user can set up the sequencing run with required fields in the SampleSheet (see example below), load consumables and kick off the run. After the sequencing run is completed, the instrument (NextSeq 2000) will auto-launch the onboard DRAGEN Single Cell RNA app using the parameter provided in the Sample Sheet. The analysis result will be saved in the same output folder of the sequencing run. For the purpose of this presentation, the “Analysis” folder was compressed and uploaded to a BaseSpace Sequence Hub project.
Table 1. Run information and sequencing mode settings.
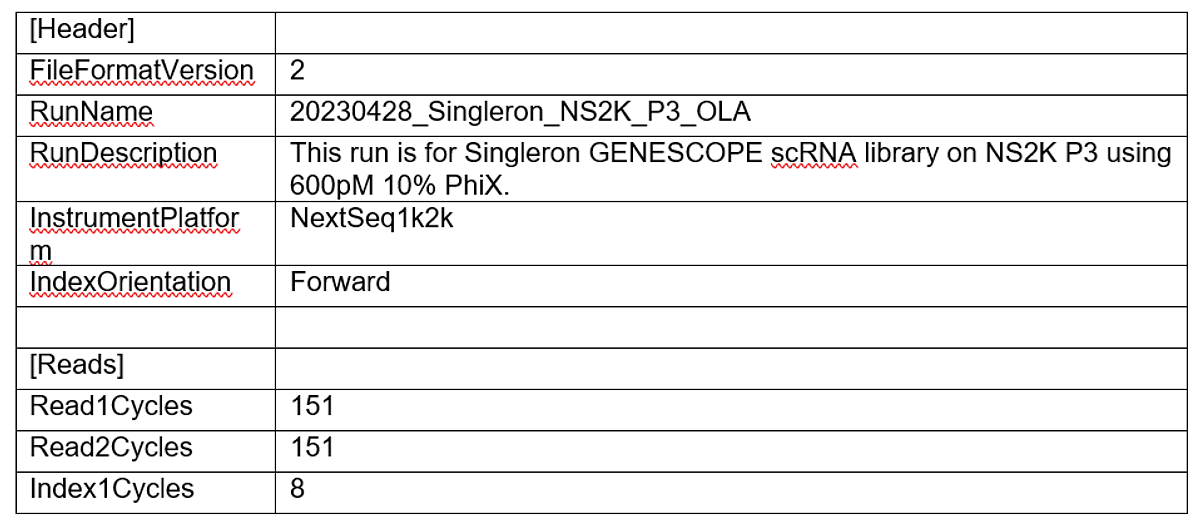
Table 2. BCLconvert setting (see user guide - https://support.illumina.com/downloads/bcl-convert-user-guide.html).
Run link: https://basespace.illumina.com/s/85WBFzC6BTEx
Project link: https://basespace.illumina.com/s/V2RDa4bJSzyp
The sequencing library was loaded at 600pM with 10% PhiX (v/v) and was sequenced with a read length of 151 + 8 +151 using the P3 flow cell. The NextSeq 2000 run meets Illumina specifications (https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000/specifications.html). We observed the same base composition and Q30 pattern as that seen from the NovaSeq 6000 run of Singleron GEXSCOPE published in our previous blogpost.
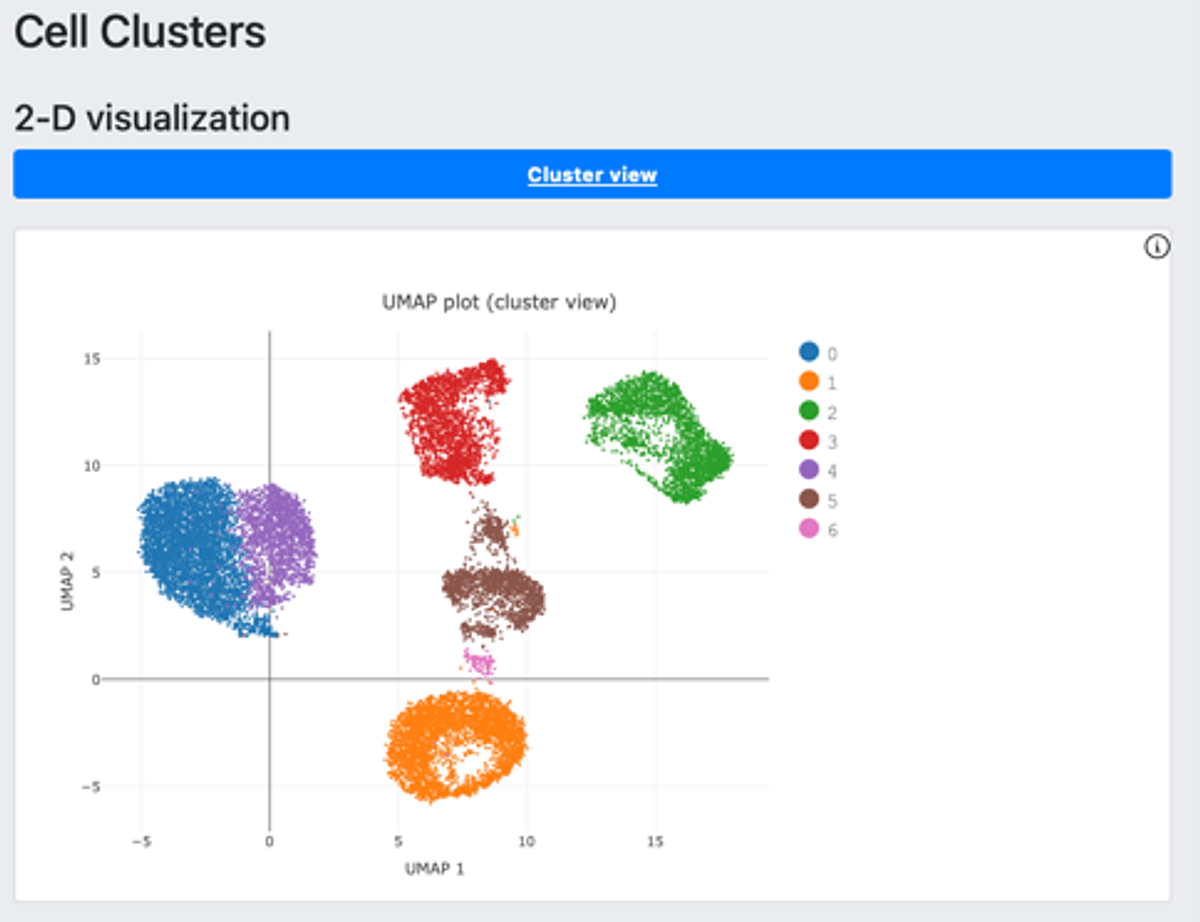
The auto launched analysis generated a DRAGEN Single Cell RNA report, for a glimpse of the analysis results. The report has three modules including cell clusters (Figure 2-A), single cell metrics (Figure 2-B) and mapping metrics (Figure 2-C).
2.A.
2.B.
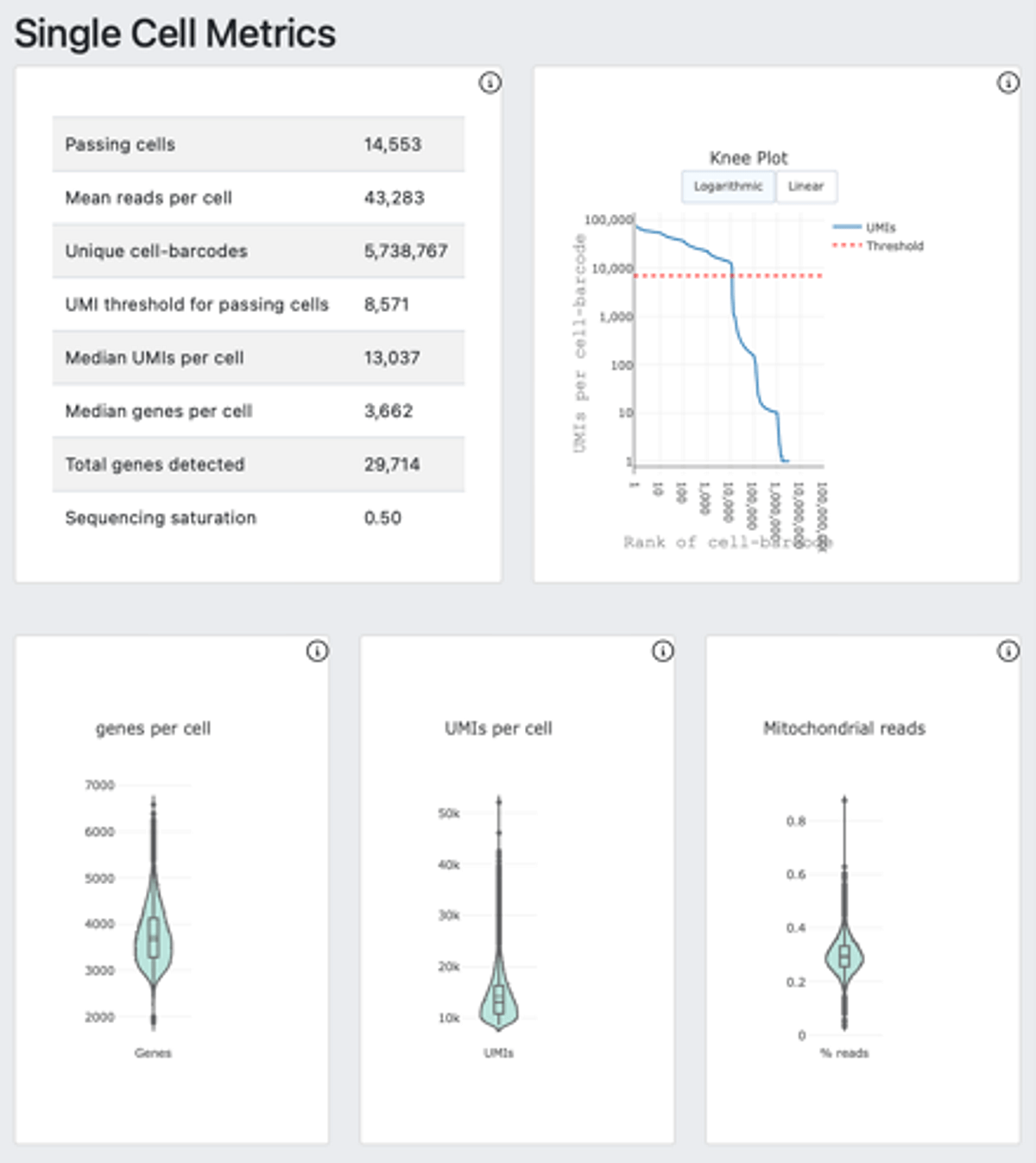
2.C.
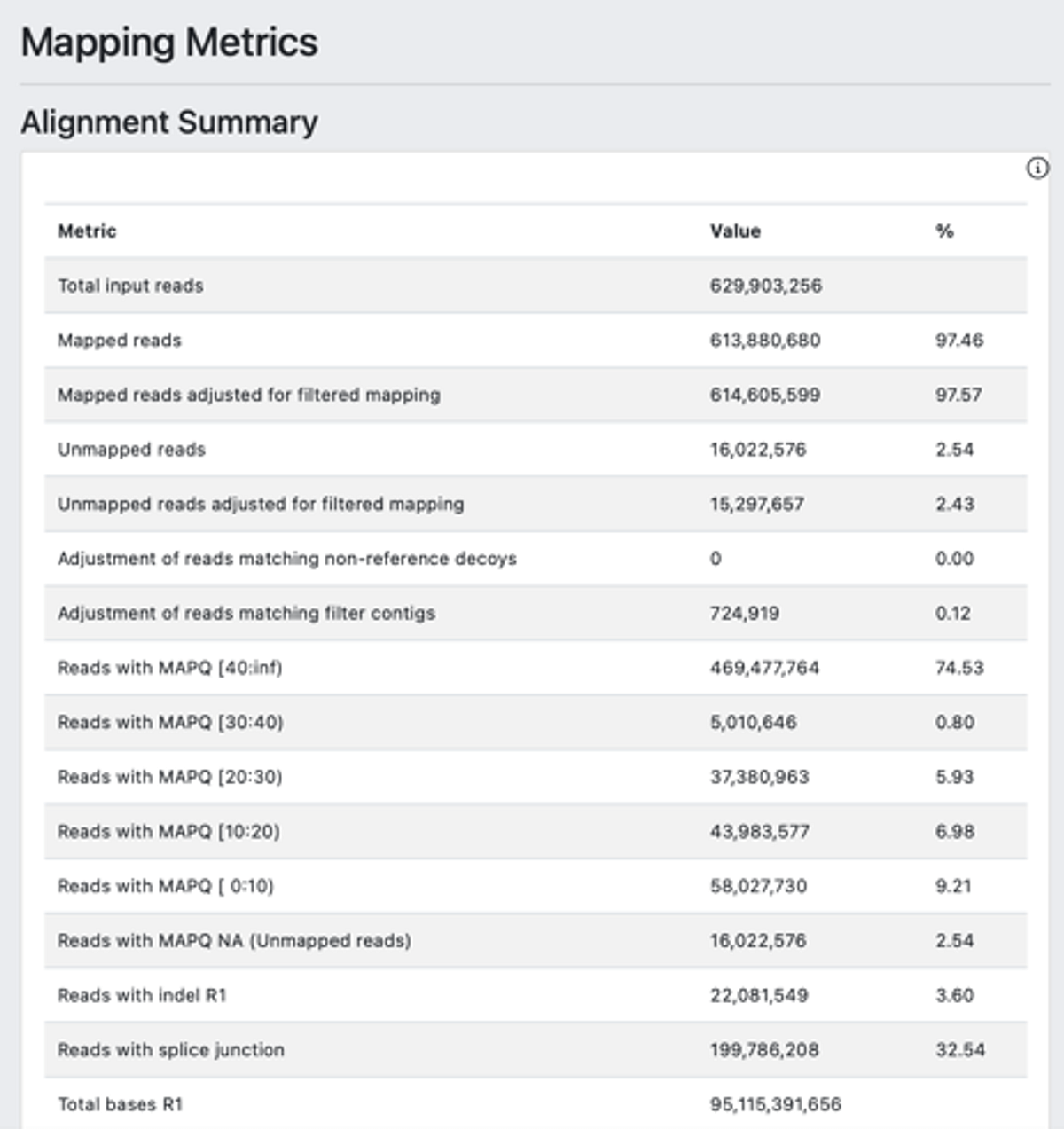
Figure 2. DRAGEN scRNA-seq offers a summary glimpse of the analysis result with cell clusters (A), single cell metrics (B) and mapping metrics (C).
Lastly, the analysis yielded a standard sparse gene expression matrix output (Figure 3) and is ready for being plugged into further tertiary analysis tool such as Seurat and Scanpy.

3.A. B0927_OLA_20230501.scRNA.barcodes.tsv
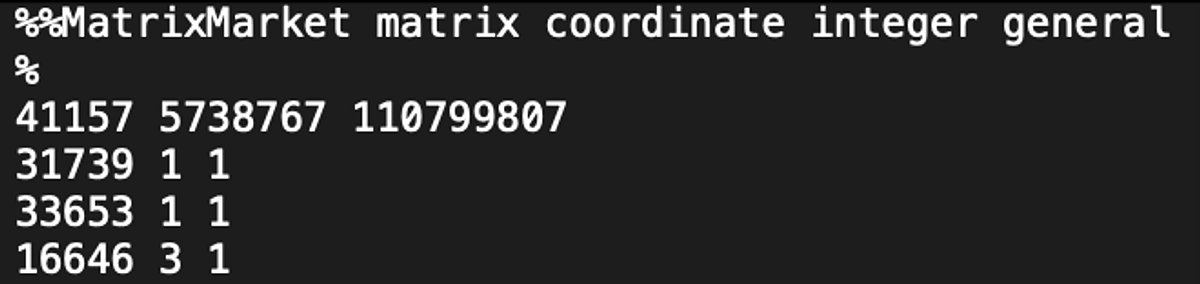
3. B. B0927_OLA_20230501.scRNA.matrix.mtx
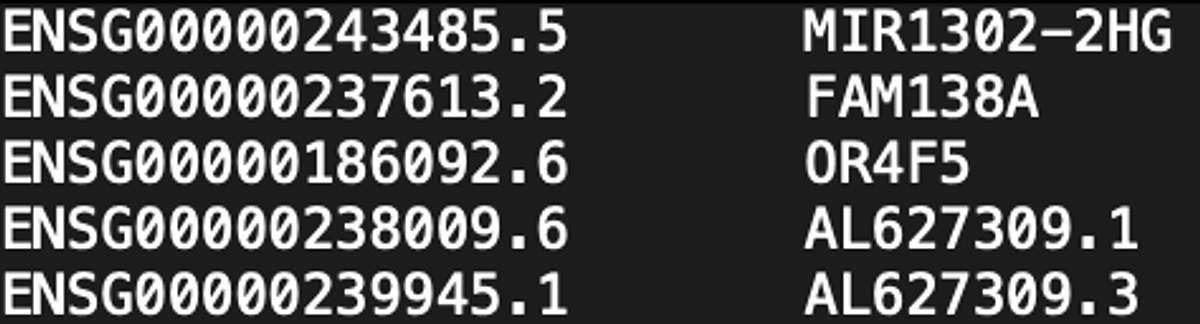
3. C. B0927_OLA_20230501.scRNA.genes.tsv
Figure 3. Snapshot of the standard sparse gene matrix files.
In conclusion, an integrated workflow from sequencing to data analysis for scRNA-seq is available on NextSeq 2000, easy to use, efficient and accurate.
Additional resources:
- https://support-docs.illumina.com/SW/DRAGEN_v40/Content/SW/DRAGEN/SingleCellRNA.htm
- https://developer.illumina.com/news-updates/singleron-gexscope-single-cell-rna-seq-data-now-available-on-illumina-basespace-sequence-hub
- https://support.illumina.com/downloads/sample-sheet-v2-template.html
- https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000/specifications.html
- https://singleron.bio/product/list-28.html
Support
Contact Singleron for any questions regarding the single cell library prep assay. Contact Illumina Tech support for sequencing related questions and DRAGEN single cell RNA pipeline.
Special thanks to our colleagues at Singleron for providing all the libraries. Hat tip to Kent Vo from the Illumina DevOps Engineering team for publishing all the data.
For Research Use Only. Not for use in diagnostic procedures. M-GL-02200