- Home
- News & Updates
- Singleron GEXSCOPE® Single Cell RNA-seq Data Now Available on Illumina® BaseSpace™ Sequence Hub
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 10/18/2022
Singleron GEXSCOPE® Single Cell RNA-seq Data Now Available on Illumina® BaseSpace™ Sequence Hub
Single cell sequencing enables studying transcriptome-wide gene expression across thousands of individual cells simultaneously, in one experiment. Singleron Biotechnologies specializes on development and commercialization of single-cell multi-omics products that can be used in both research and clinical settings. Singleron GEXSCOPE Single Cell RNA Library Kit is a complete solution for transforming tissues into single cells and converting the mRNA contained in the single cells into next-generation sequencing (NGS) libraries.
Singleron GEXSCOPE single cell RNAseq workflow
Singleron GEXSCOPE workflow starts with dissociation of tissues into single cell or single nuclei suspensions. In the next step, Singleron SCOPE-chip®, a portable, microfluidic chip with microwells, integrates multiple steps of the processing workflow such as single cell partitioning, cell lysis and capture of cellular mRNA (Figure 1). The SCOPE-chip is easy to use and can be operated manually without special equipment.
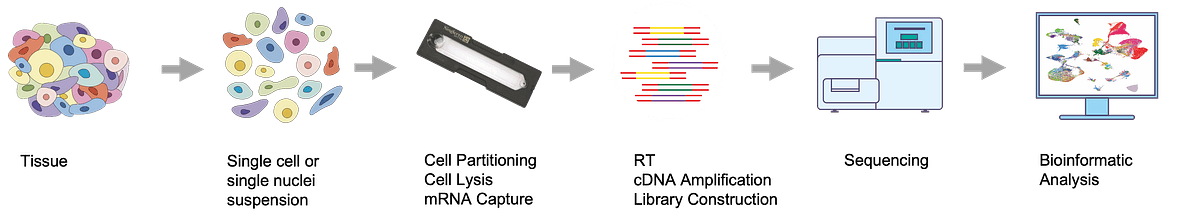
Figure 1 – Experimental workflow
Captured mRNA is subjected to reverse transcription and barcoded with unique cell barcodes, followed by NGS library preparation which is compatible with all Illumina sequencing platforms but the NovaSeq™ 6000 is recommended due to the necessary output. Singleron recommends 50,000 reads per single cell. No custom sequencing primers is required.
The obtained paired-end FASTQ files can be processed with Singleron CeleScope® pipeline that enables expression matrix generation, clustering and marker gene expression analysis.
Singleron GEXSCOPE single cell RNAseq run available on BaseSpace™ Sequence Hub
To view Singleron GEXSCOPE single cell RNAseq run on NovaSeq 6000 v1.5 S4 flow cells, please access the BaseSpace Sequence Hub and import the sequencing run using the link provided below. Singleron GEXSCOPE single cell RNAseq can be found on the lane 3 and 4 of the flowcell.
These demo data are free to use on the BaseSpace platform (see our previous post) and can be used for direct comparison with your own single cell sequencing data generated with GEXSCOPE Single Cell RNA Library Kit. To understand how to evaluate your sequencing run, see our post: "Does my sequencing run look good"?
Singleron GEXSCOPE single cell RNAseq libraries were sequenced with a read length of 151 + 8 + 8 +151, and 4% PhiX (v/v) was spiked in the pool of libraries. Because of the position of polyT (starting from ~70bp in R1 read), the % base after 70bp in read 1 is unbalanced, and the sequencing quality drops. This is an expected phenotype.
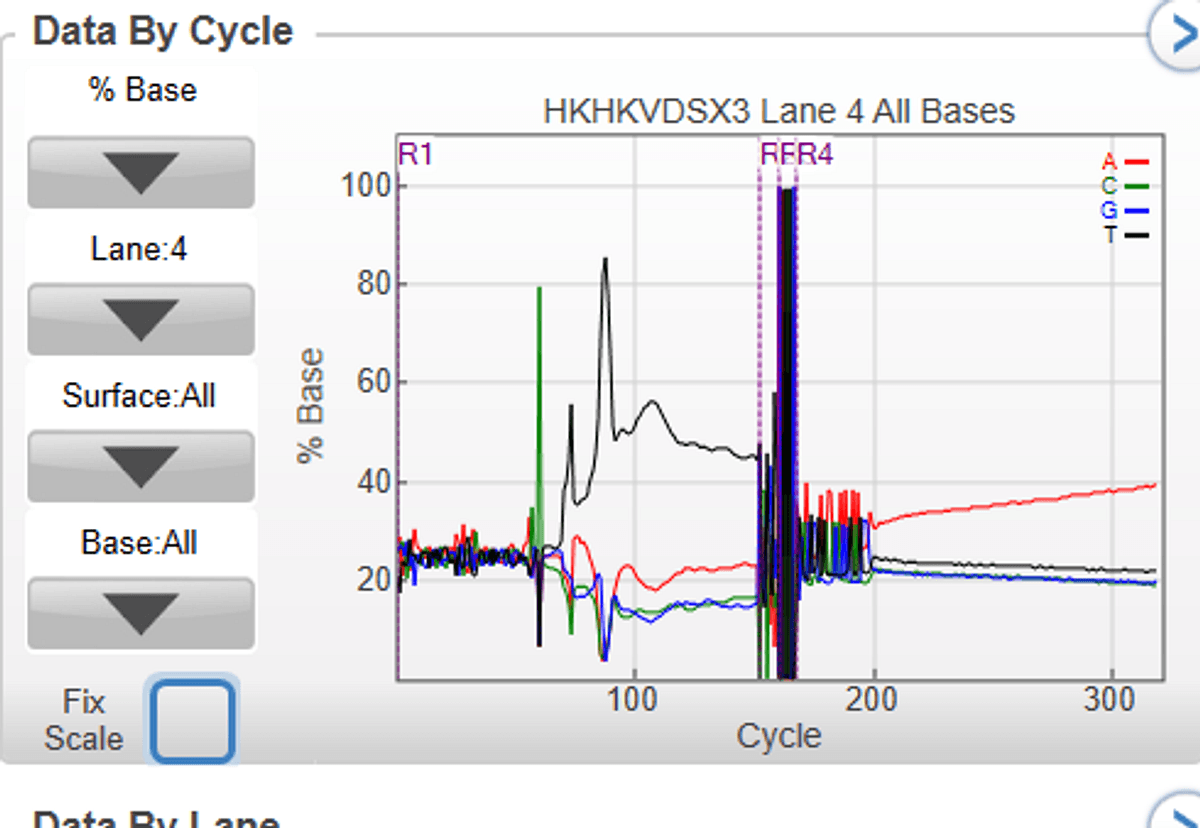
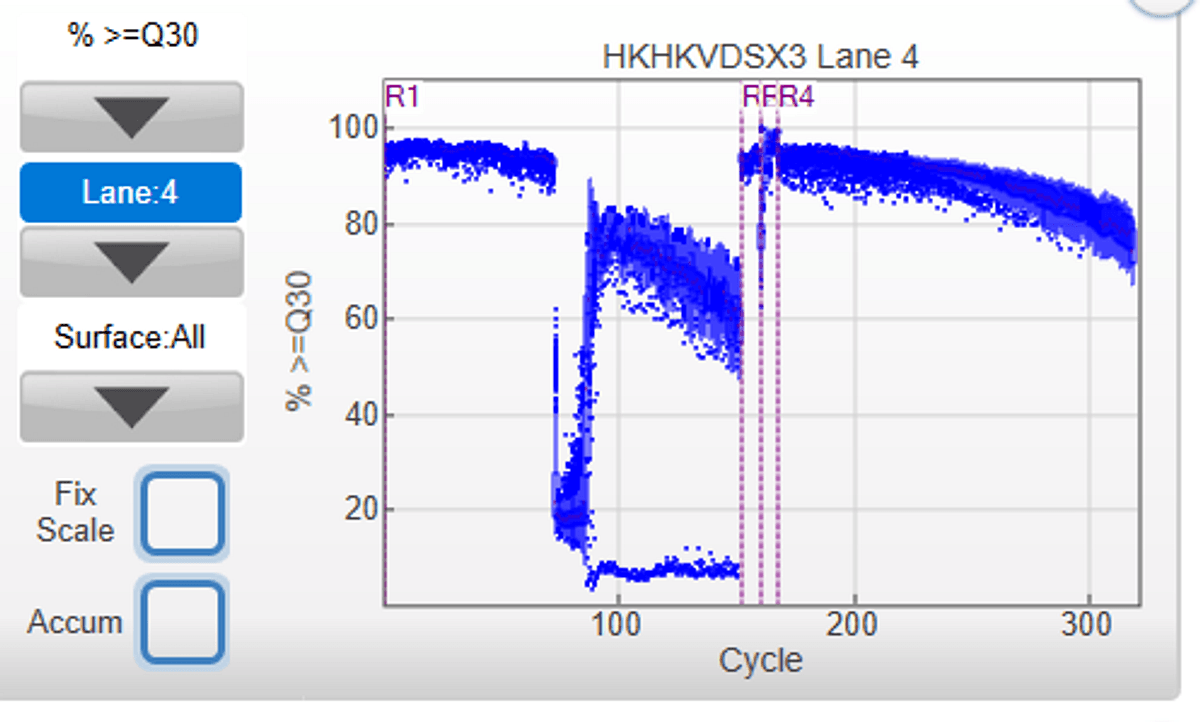
Link to run: https://ilmn-sso.basespace.illumina.com/s/sdiZ5I3HQLdt
Link to project (contains the Celescope report): https://ilmn-sso.basespace.illumina.com/s/gabUz99LeXhG
Additional resources:
To answer your questions regarding preparation and sequencing of GEXSCOPE libraries, contact our experts at Singleron (info@singleronbio.com). For questions about your sequencing run, contactIllumina (techsupport@illumina.com).
If you need further information on Singleron’s single cell sequencing technology, kits and instruments, visit Singleron’s product page.
For Research Use Only. Not for use in diagnostic procedures.
Special thanks to our colleagues at Singleron for sharing sequencing run data and dataset generated with the CeleScope® pipeline.