- Home
- News & Updates
- Qiagen QIAseq Immune Repertoire RNA Library Kit data on the NextSeq™ 2000 – 600 cycles kit are now available on BaseSpace™ Sequence Hub!
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 04/07/2023
Qiagen QIAseq Immune Repertoire RNA Library Kit data on the NextSeq™ 2000 – 600 cycles kit are now available on BaseSpace™ Sequence Hub!
Accurate characterization of the T cell receptor (TCR) repertoire is key to understanding adaptive immune responses and has applications across vaccine development, autoimmunity, monitoring treatment response in lymphoid malignancies and immunotherapy. Compared to traditional methods, next-generation sequencing (NGS) provides an unprecedented, high-resolution picture of the immune repertoire. However, the complex workflows and high time investment required can make developing automated NGS workflows challenging. In addition, this approach involves multiplex PCR with primers targeting different V or J regions, which can introduce substantial amplification bias.
The QIAseq Immune Repertoire RNA Library Kits from Qiagen use Unique Molecular Indices (UMI) with gene-specific primers to target the T-cell receptor α, β, γ, and δ genes (TRA, TRB, TRG, TRD) for NGS sequencing. Each unique panel is carefully designed and laboratory-verified for sequencing performance with a UMI-aware alignment software for maximum sequencing performance and accurate results. The Human and Mouse T-cell Receptors Panel is used for sequencing the recombined, full-length V(D)J region of the α, β, γ, and δ genes, including the CDR3 regions. Online analysis through the Qiagen GeneGlobe Data Analysis Center provides key sequencing QC metrics, as well as the frequency and identity of each clonotype sequenced.
Today, we are excited to present NextSeq 2000 data on the newly launched 600 cycles sequencing kits with the QIAseq Immune Repertoire RNA Library Kits.
Sample, library, and sequencing preparation
Human whole blood from a single healthy subject was obtained from AllCells. Peripheral blood mononuclear cells (PBMCs) were isolated using SepMate PBMC Isolation Tubes (STEMCELL Technologies; 85415). Total RNA was isolated from multiple PBMC aliquots using the RNeasy Plus Mini Kit (Qiagen; 74134). A total of six libraries were constructed in duplicate using 20ng, 50ng, and 100ng RNA inputs with the QIAseq Human TCR Panel Immune Repertoire RNA Library Kit following manufacturer’s recommendations (Qiagen; 333705). Final dual-indexed libraries were validated using the High Sensitivity D1000 ScreenTape Assay for TapeStation (Agilent; 5067-5584) and Qubit dsDNA HS Kit (Q32851) prior to library pooling. Custom primers were spiked into corresponding reagent reservoirs following Illumina’s recommendations. The library pool was spiked with 10% Phix and diluted to 450pM prior to sequencing with the NextSeq 1000/2000 P1 Reagents (600 Cycles) catalog number 20075294.
Accessing sequencing run
NextSeq 1000/2000 sequencing data from the QIAseq Immune Repertoire RNA Library Kits libraries were automatically streamed to BaseSpace Sequence Hub platform where pooled sample data is automatically de-multiplexed into separately dual-indexed library sequencing information. To view this run on NextSeq2000 600 cycles kit, please access the BaseSpace Sequence Hub and import the sequencing run and FASTQ files information using the links provided below.
Run link: https://basespace.illumina.com/s/louBJbc1xoSC
Project link: https://basespace.illumina.com/s/FAbkwrYIWMvo
These demo data are free to use on the BaseSpace Sequence Hub platform (see our previous post - https://developer.illumina.com/news-updates/demo-data-on-illumina-basespace-sequence-hub)
Evaluation of the sequencing run
To understand how to evaluate your sequencing run, see our previous blogpost "Does my sequencing run look good?"
QIAseq Immune Repertoire RNA Library Kits libraries were sequenced with a read length of 301 + 8 + 8 + 301 on the NextSeq1000/2000 P1 100M Reagents (600 Cycles), catalog number 20075294. With over 127M Reads Passing Filters observed at average %>Q30 above 86% across the run, this dataset not only exceeds specifications for the NextSeq 1000/2000 P1 100M Reagent Kit, but also clearly demonstrates the premiere performance of the NextSeq 1000/2000 600 cycles Kits for obtaining full-length TCR V(D)J receptor information for α, β, γ, and δ genes using the QIAseq Immune Repertoire RNA Library Kits. (Figure 1).
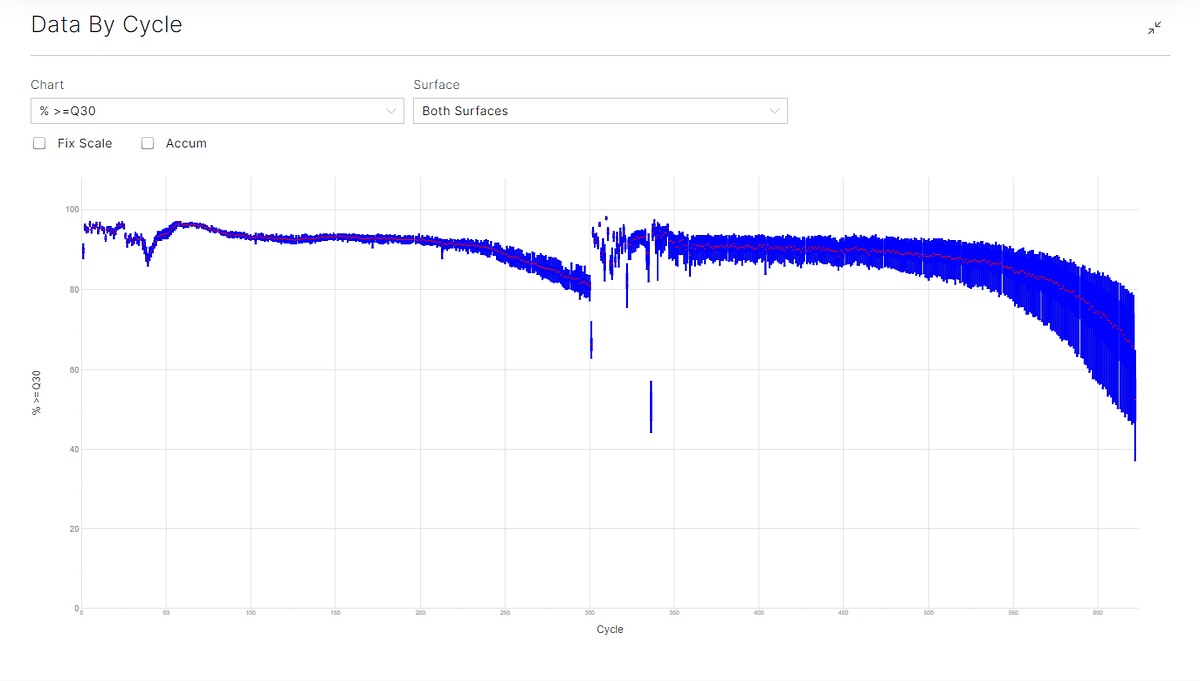
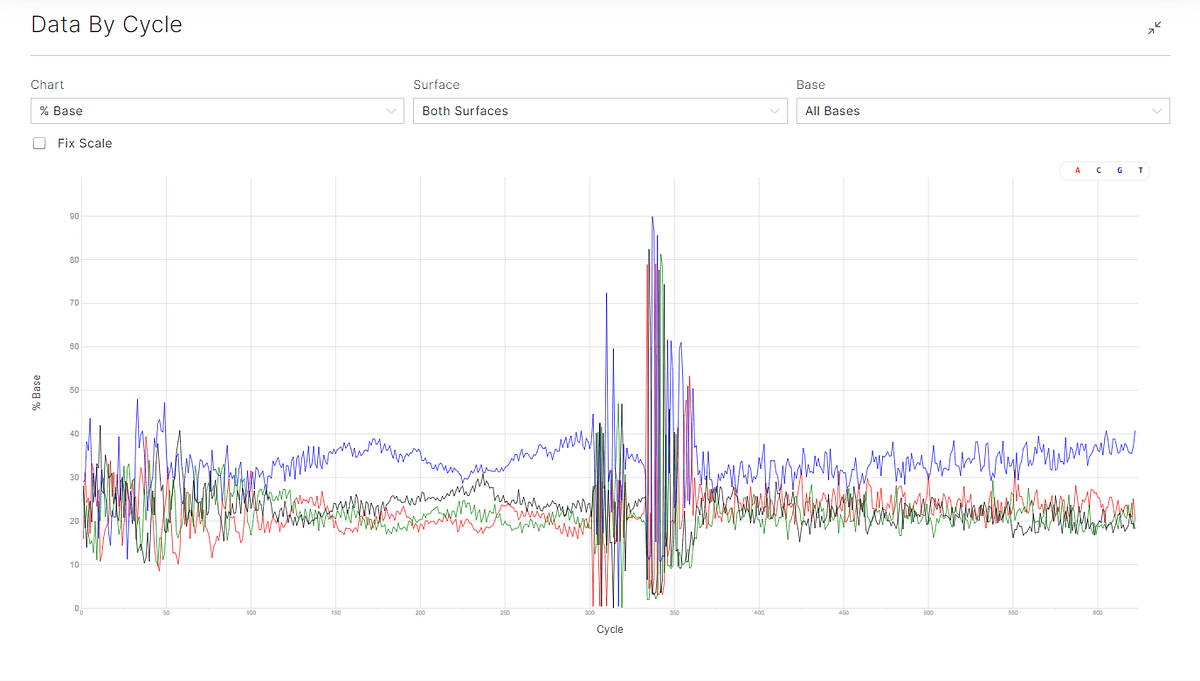
Figure 1: sequencing data by cycle showing the data by cycle for Q30 (top), and percentage base by cycle (bottom).
Analysis
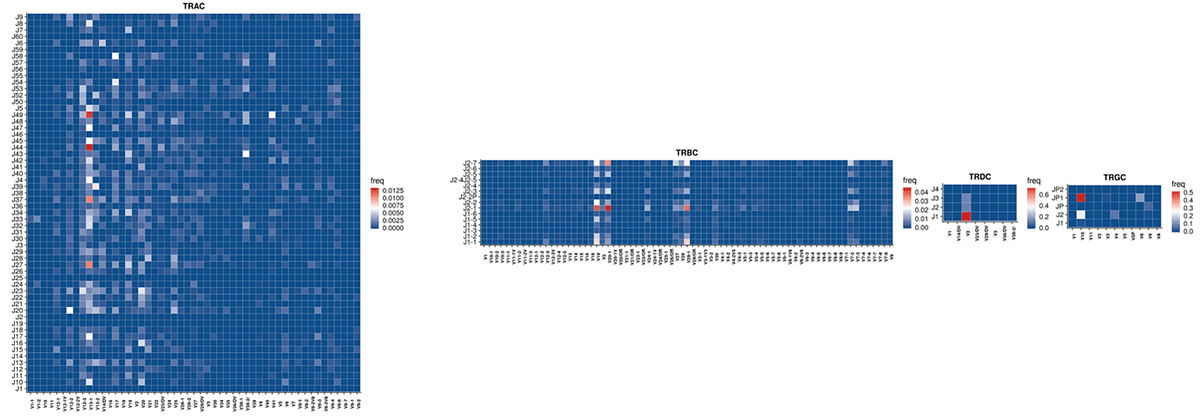
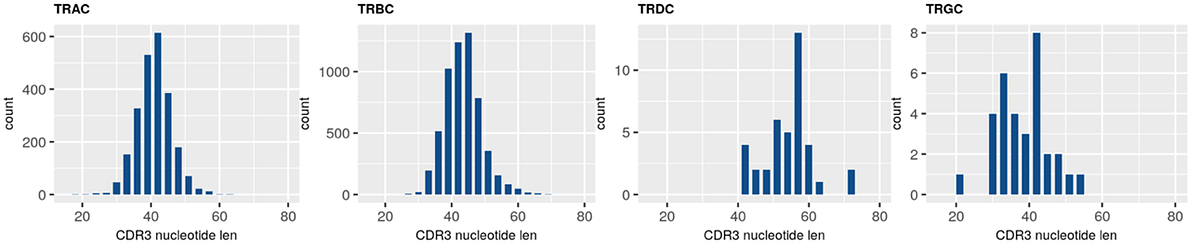
Raw sequencing data was automatically streamed from the NextSeq 2000 instrument to BaseSpace Sequence Hub where FASTQ information was automatically generated. Using the Qiagen GeneGlobe Data Analysis Center, FASTQ files were directly imported from BaseSpace Sequence Hub to GeneGlobe where they were analyzed using IMSeq with modifications to enable UMI directed error-correction based on multiple reads of the same molecules. Multiple table and graphical outputs were automatically generated providing a comprehensive summary of UMI-error correction, clonotype frequencies, and repertoire metrics highlighted in Figure 2.
Resulting secondary clonotype analysis and repertoire features analysis concludes consistent and expected results across all replicates at varying RNA inputs. Highly consistent UMI and clonotype counts were observed across replicates indicating the high robustness of the assay with UMI error correction accuracy. Increased clonotype counts were well correlated with libraries input with increasing RNA amounts as seen in the rarefaction analysis. Consistent V(D)J gene usage was also observed across replicates and is in line with expectations for features of a normal healthy TCR repertoire.
A. V-J usage heatmap for sample “Qiagen-100ng-1”
B. CDR3 length plot for sample “Qiagen-100ng-1”
C. Rarefaction plot
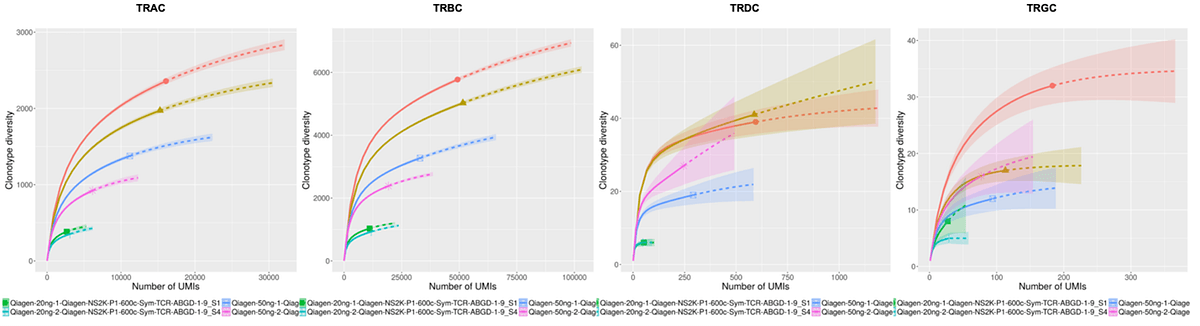
Figure2. Plots of IR features generated on the Qiagen GeneGlobe Data Analysis Center: A. V-J usage heatmap B. CDR3 length C. Rarefaction plot
Table1. A summary from read pairs to clonotypes counts across all samples.
Metric |
Qiagen-20ng-1 |
Qiagen-20ng-2 |
Qiagen-50ng-1 |
Qiagen-50ng-2 |
Qiagen-100ng-1 |
Qiagen-100ng-2 |
read pairs total |
17,339,192 |
17,531,473 |
17,080,772 |
17,957,882 |
18,225,347 |
15,367,793 |
UMI consensus read pairs usable as input to IMSEQ |
24,479 |
25,736 |
89,141 |
49,334 |
132,224 |
126,546 |
UMI consensus read pairs usable for clonotype calls |
13,792 |
15,010 |
44,565 |
25,741 |
66,111 |
67,549 |
UMI consensus read pairs usable for clonotype calls TRAC, percent |
38.90% |
45.80% |
42.80% |
40.50% |
42.00% |
37.60% |
UMI consensus read pairs usable for clonotype calls TRBC, percent |
81.60% |
78.20% |
75.40% |
76.60% |
75.30% |
74.90% |
UMI consensus read pairs usable for clonotype calls TRDC, percent |
12.10% |
7.90% |
14.00% |
16.80% |
16.70% |
16.50% |
UMI consensus read pairs usable for clonotype calls TRGC, percent |
0.70% |
0.90% |
0.50% |
1.00% |
0.70% |
0.80% |
clonotypes TRAC |
384 |
353 |
1,376 |
922 |
2,358 |
1,972 |
clonotypes TRBC |
1,032 |
927 |
3,272 |
2,383 |
5,771 |
5,025 |
clonotypes TRDC |
6 |
6 |
19 |
27 |
39 |
41 |
clonotypes TRGC |
8 |
5 |
12 |
16 |
32 |
17 |
Html reports have also been uploaded on the BaseSpace project: https://basespace.illumina.com/s/FAbkwrYIWMvo
Learn more:
https://geneglobe.qiagen.com/us/analyze
https://www.illumina.com/syste...
https://www.illumina.com/systems/sequencing-platforms/nextseq-1000-2000/specifications.html
https://www.illumina.com/areas-of-interest/immunogenomics.html
Support contact information:
To answer your questions regarding preparation and sequencing of Qiagen immune repertoire libraries, contact Qiagen (https://www.qiagen.com/no/know...).
For questions about your sequencing run, contact Illumina (techsupport@illumina.com).
Special thanks to our colleagues at Qiagen for providing library prep reagents and controls for this experiment and analyzing the IR-data. Hat tip to Illumina Scientists Robin Bombardi and Qiyuan Han and the Emerging Apps team for sequencing these runs for this collaboration.
For Research Use Only. Not for use in diagnostic procedures. M-GL-01592