- Home
- News & Updates
- Spatial transcriptomics data from Curio Bioscience is now available on BaseSpace™ Sequence Hub
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 03/14/2024
Spatial transcriptomics data from Curio Bioscience is now available on BaseSpace™ Sequence Hub
We are excited to announce the availability of NextSeq™ 2000 data on BaseSpace Sequence Hub from Curio Bioscience, a partner that develops high-precision tools for the spatial biology market.
Next-generation sequencing is an ideal tool for high-throughput discovery and translational spatial transcriptomics studies due to its general accessibility, ease of use, throughput capabilities, and ability to measure transcripts with high specificity.
The Curio Seeker 3x3 Spatial Mapping Kit is a high-throughput spatial sequencing platform that enables whole transcriptome analysis of a 3 mm x 3 mm section of fresh frozen tissue at single-cell scale resolution. The Curio Seeker 3x3 Spatial Mapping Kit’s workflow is amenable to a wide range of tissue types and organisms, requires no optimization, and makes use of standard molecular biology reagents and equipment. No new capital equipment, specialized personnel, or custom gene panels are required.
The Curio Seeker 3x3 tile, bead structure, and workflow used to generate the presently announced data are illustrated in Figures 1 and 2 below.
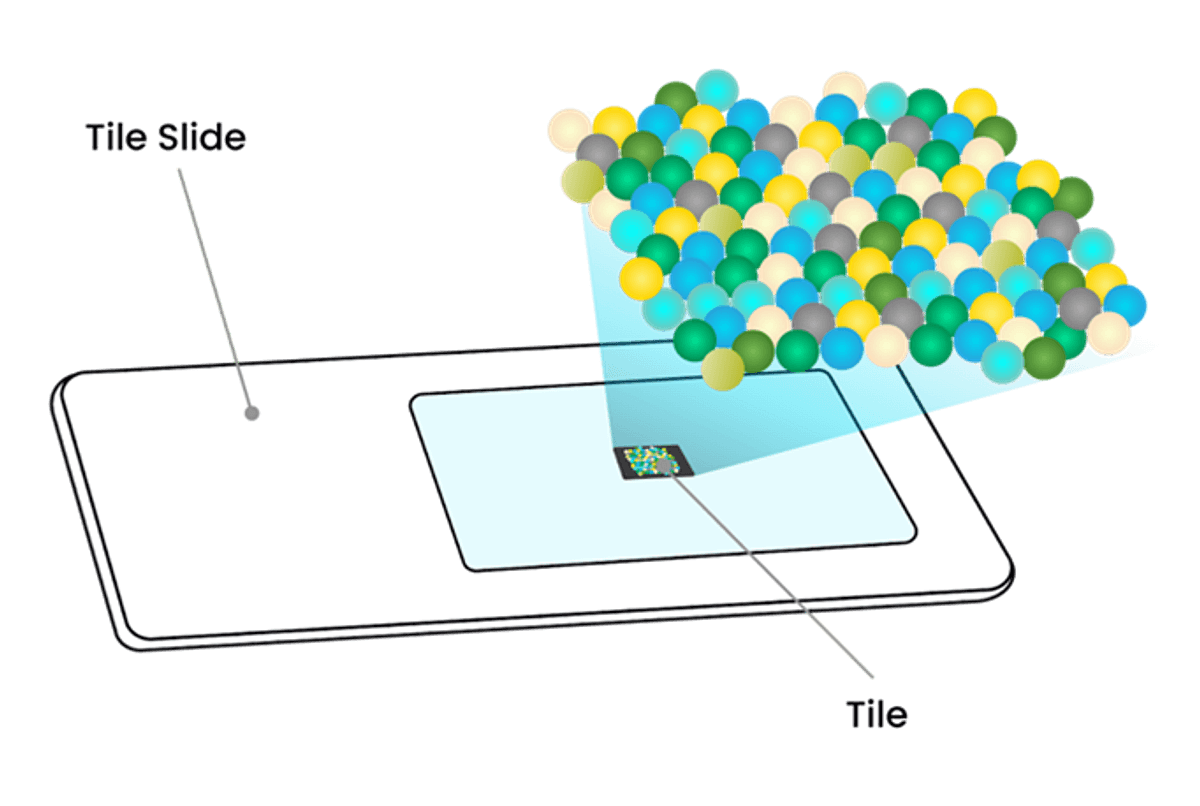
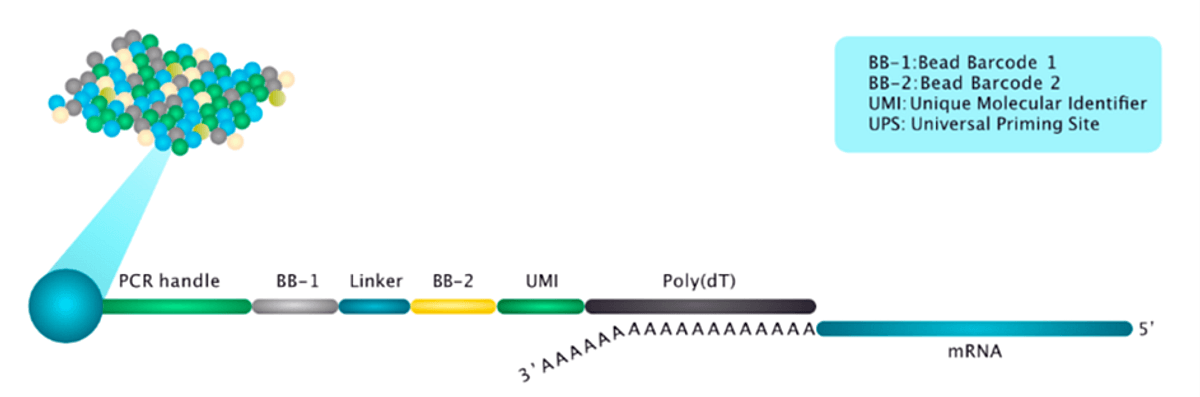
Figure 1: The Curio Seeker 3x3 tile (top figure) and bead structure (bottom figure)

Figure 2: The Curio Seeker workflow.
According to Curio, the workflow was completed in 8 hours with 2.5 hours of hands-on time and multiple safe stopping points along the way. A 10 micron section of fresh frozen mouse brain was melted onto the Curio Seeker 3x3 tile and placed in hybridization buffer, where the polyA tails of the tissue mRNA were annealed to the polydT capture sites on the beads. Once hybridized, reverse transcription polymerized the first strand cDNA to the bead and barcode. The beads and tissue were then cleared from the glass support, and the beads underwent second strand synthesis and cDNA amplification with the reagents in the Curio Seeker 3x3 Spatial Mapping Kit. The resulting cDNA was tagmented by the Nextera™ XT kit and indexed with reagents from the Curio Seeker Dual Indexing Primers Kit V2. Once cleaned up, the resulting library was sequenced on the Illumina NextSeq 2000 and could have been sequenced on the NextSeq 1000, NovaSeq™ X, and NovaSeq 6000 sequencers as well.
Sequencing depth for each experiment depends upon experimental aims. For shallow sequencing to check library quality and preview spatial expression, Curio recommends ~30M reads per tile. For deep sequencing, Curio recommends 200M-600M reads per tile depending on the specific tissue type and tile coverage. How many samples run on each flowcell will depend on the depth desired. For this blogpost, we are showcasing NextSeq 2000 run data with the Curio Seeker libraries.
Sequencing Run on NextSeq 1000 / 2000 Illumina platform.
Below are two links to directly import the runs and project folders into your BaseSpace account. These runs can be found under the “spatial omics” and “RNA-seq” category. Because these are public data sets (https://basespace.illumina.com...), these are free and do not count against storage limits. See our blogpost to understand how to access these data.
You can use the demo data to compare with your own single cell runs. See our first blog post in this series for additional details on how to evaluate your sequencing run quality. A high quality Q30 score is maintained on NextSeq 2000 (Figure 3) with Curio libraries. Please see the Curio Seeker 3x3 Spatial Mapping Kit User Guide for more information on how to sequence these libraries.
NextSeq 2000 run link: https://ilmn-sso.basespace.illumina.com/s/GbwA03sgTUXq
Project link: https://ilmn-sso.basespace.illumina.com/s/Zg7cVyBX7pJU
Categories: spatial omics, RNA-seq
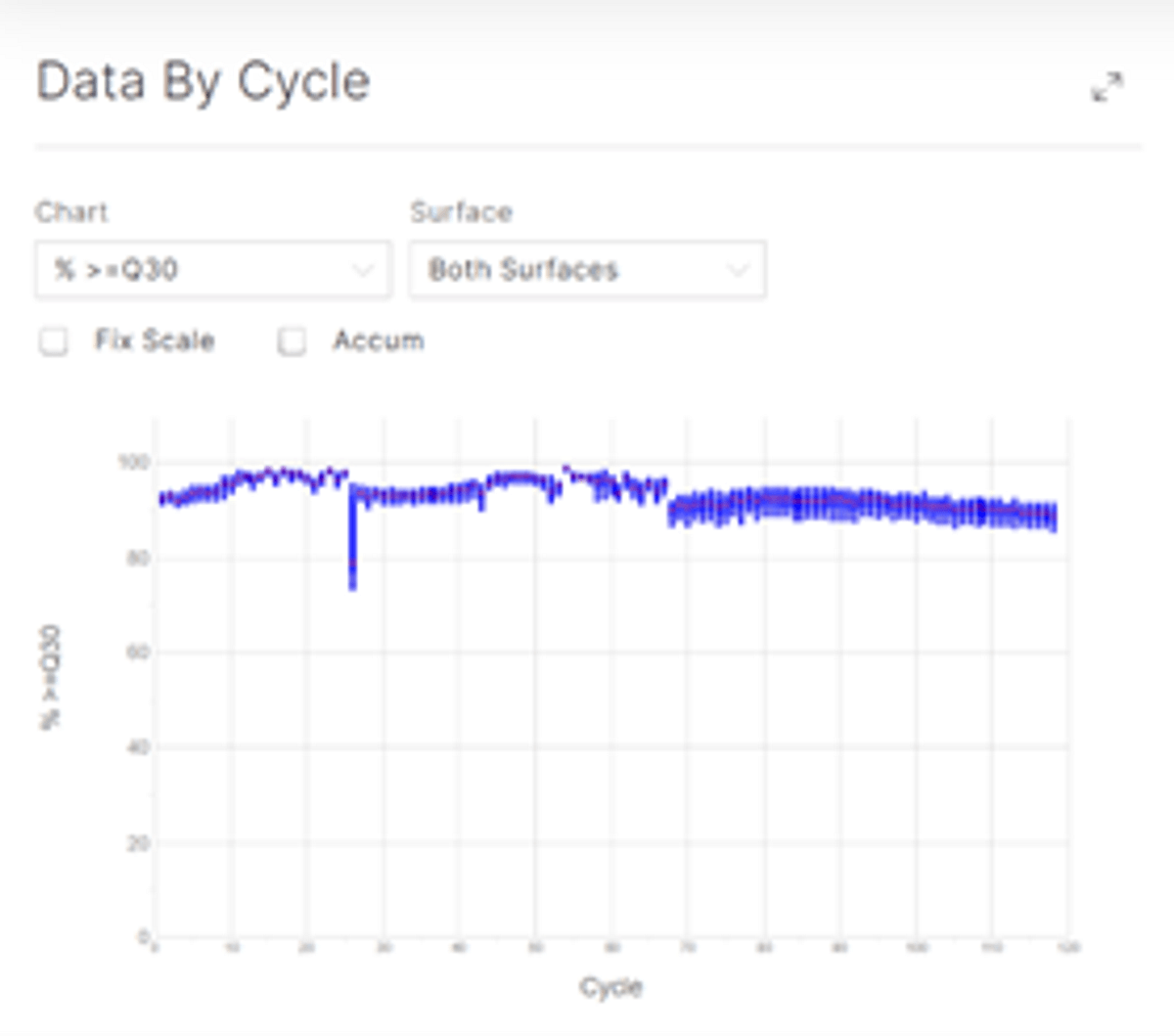
Figure 3: A high quality Q30 score is maintained on NextSeq 2000.
Curio Seeker pipeline access
Untrimmed FASTQ files from the sequencing library are available in the BaseSpace Project and uploaded to the Curio Seeker bioinformatics pipeline. You can access the pipeline through the Curio Bioscience bioinformatics portal or download the pipeline for internal processing. Please contact our techsupport@curiobioscience.com to access the portal or download the pipeline. Once analyzed, the pipeline provides a variety of bead-by-gene matrices that plug into different secondary analysis platforms and a sample report, which gives researchers a glimpse of their tile run and helps them determine next steps.
Curio Seeker sample report summary
FASTQ files from the mouse hippocampus sample, and a pdf of the same report generated by the Curio Seeker bioinformatics pipeline can now be accessed from the BSSH demo page, under the Project.
The sample report provides metrics to determine the quality of your run, if you have captured your region of interest, an unbiased UMAP cluster plot and the spatial transcriptomics image of the region captured on the tile.
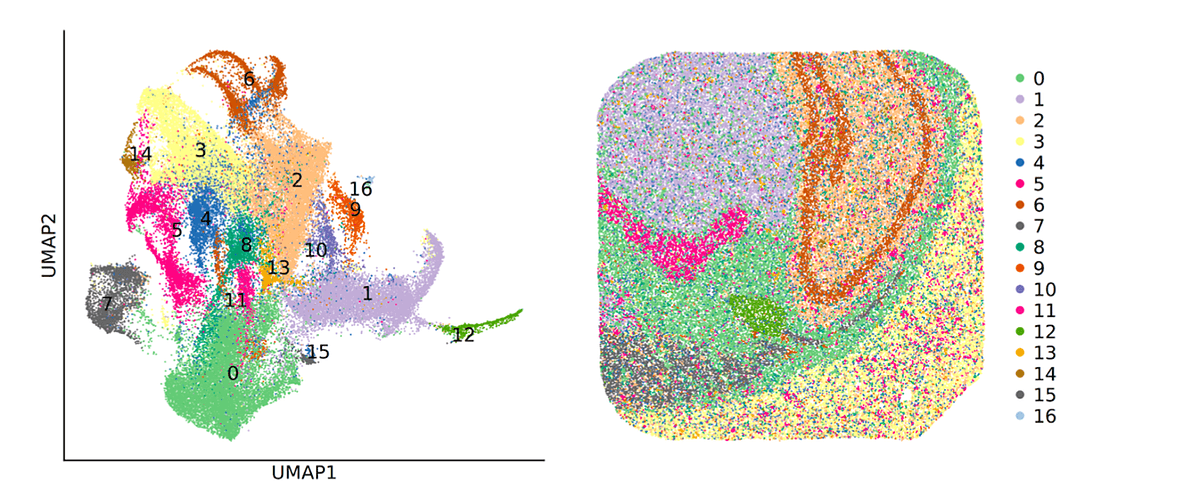
Figure 4: Unbiased UMAP cluster plot and spatial transcriptomic map. Dimension reduction, unbiased clustering, and spatial transcriptomics map. Standard html output from the Curio Seeker bioinformatics pipeline includes results from dimension reduction and unbiased clustering (left) as well as spatial map of identified clusters (right).
Support
To learn more about the Curio Seeker 3x3 Spatial Mapping Kit, please visit us at www.curiobioscience.com or contact us at techsupport@curiobioscience.com. Contact Illumina Tech support for sequencing related questions.
Special thanks to our colleagues at Curio Bioscience for providing the data.
This material is provided for informational purposes only, to provide an example of the range of capabilities offered by Illumina’s innovative instruments and technologies relating to the analysis of genetic variation and function. It is not intended to be, and should not be construed as, an endorsement of specific third-party products, technologies, or services. Any data presented is neither owned nor verified by Illumina. Customers should independently determine whether using any third-party solutions on Illumina platforms is appropriate for their workflow or laboratory.
For Research Use Only. Not for use in diagnostic procedures. M-GL-02599