- Home
- News & Updates
- Illumina Connected Multiomics now in full customer release
-
Illumina Connected Multiomics
-
News
- 01/06/2026
Illumina Connected Multiomics now in full customer release
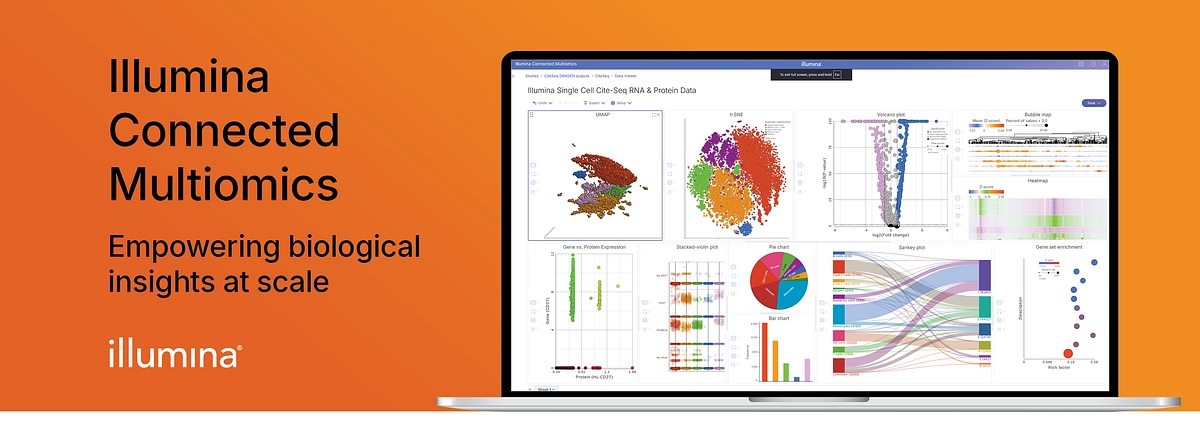
Despite the transformative potential of multiomics—integrating proteomics, transcriptomics, epigenomics, and genomics across modalities—realizing its promise is challenging. Scientists have long been forced to cobble together disconnected datasets and tools that are each optimized for a single omic layer. The result? Missed connections, shallow interpretations, and insights that remain just out of reach. But that’s changing.
With the official release of Illumina Connected Multiomics, we’re removing the barriers that have long stood in the way of discovery. Illumina’s newest cloud-based software unifies data exploration, visualization, and interpretation in one intuitive environment, enabling researchers to gain true multiomic insights, seamlessly.
Deeper insights made possible
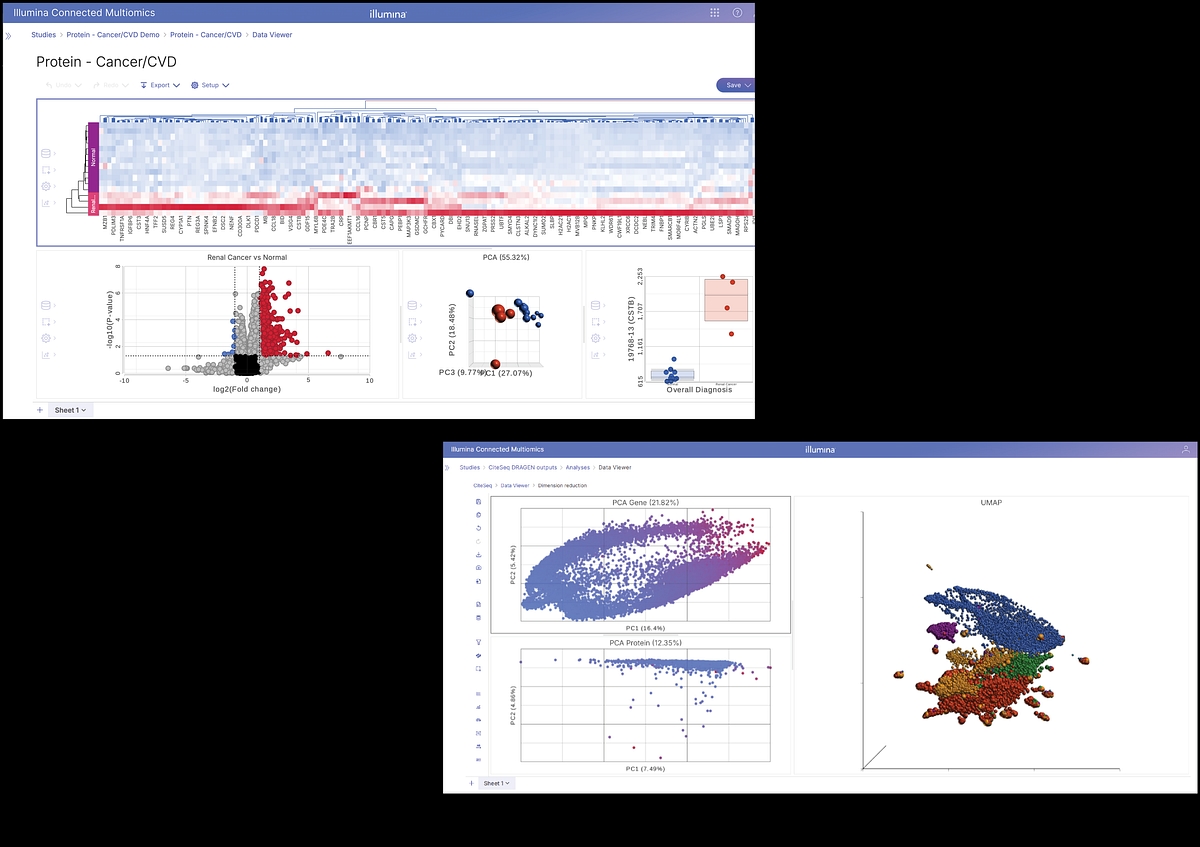
Multiomics enables scientists to explore the full central dogma of biology by integrating data from proteomic, transcriptomic, epigenomic, and genomic layers. It is a unified, cloud-based software that automates data integration, enables complex analysis, and provides rich visualization. It supports Illumina and select third-party assays, enabling seamless workflows from sample prep to interpretation.
Hosted on the AWS cloud, Connected Multiomics elastically scales with your study size and meets industry security standards. Researchers of all skill levels can independently explore multiomic datasets using an intuitive graphical interface, interactive visualizations, and robust statistical methods.
Connected Multiomics v1.0 allows you to easily analyze single omic and multiomic studies and reveal insights, including:
- Detect regulatory mutations that alter methylation patterns and influence gene expression.
- Reveal post-transcriptional regulation and its impact on protein composition.
- Uncover how proteins drive cellular functions and disease by revealing expression patterns, interactions, and post-translational modifications that shape biological pathways.
- Expose gene expression variability and signaling differences at the individual cell level, providing a high-resolution view of cellular states and interactions with single-cell analysis.
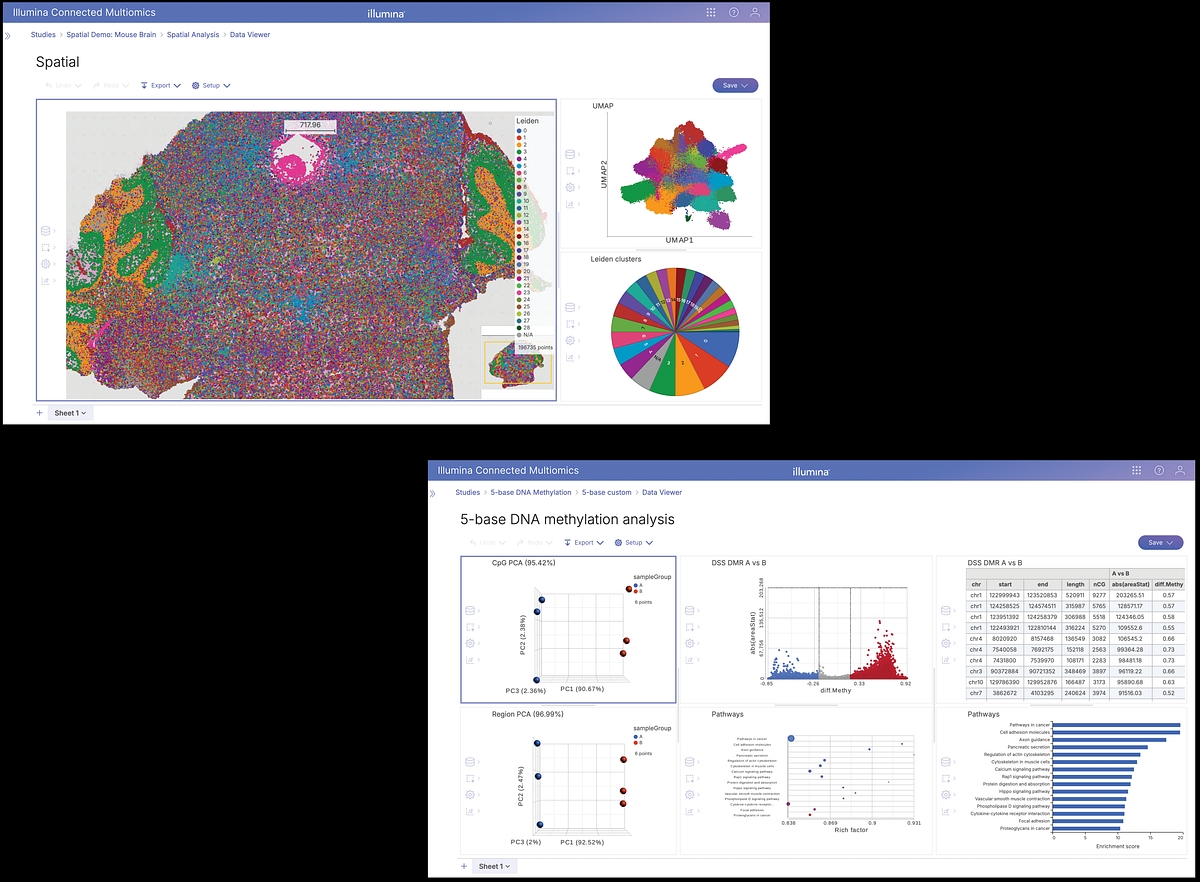
- Understand cellular interaction within native tissue context by uncovering spatial patterns that drive development, disease progression, and therapeutic response.
- Reveal a comprehensive view of epigenetic regulation using 5-base data that profiles DNA methylation at single-nucleotide level, uncovering mechanisms that drive gene expression and disease progression.
Future versions of Connected Multiomics will bring even more multiomic integration capabilities.
A Connected workflow
As part of the Illumina software environment, Connected Multiomics leverages the speed and accuracy* of DRAGEN™ secondary analysis and integrates Correlation Engine, one of the world’s largest omics knowledgebases, for biological interpretation. Illumina sequencers and key Illumina assays, including Illumina Protein Prep, 5-Base DNA Prep, 5-Base DNA Prep with Enrichment, Single Cell 3’ RNA Prep kit, and spatial transcriptomics technology** are supported through a growing number of pre-built and customizable workflows. Maximizing flexibility, Connected Multiomics also supports select third-party assays and tools, including SomaLogic, 10X Genomics, and Parse Biosciences. Find the full list of currently supported assays on our user documentation. The addition of Connected Multiomics brings exploratory analysis and statistical visualization capabilities to the Illumina multiomics pipeline, making it a complete end-to-end analysis solution.
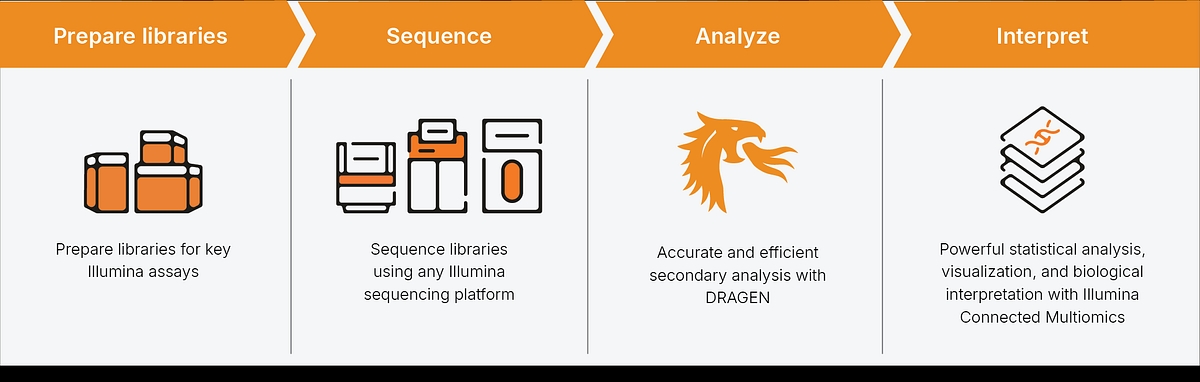
Secondary analysis
DRAGEN provides pre-configured, high-performance analysis pipelines for secondary analysis that are highly accurate. It supports Illumina and other compatible assays with powerful multiomic pipelines that integrate with Connected Multiomics to simplify complex workflows across proteomics, transcriptomics, 5-base methylation, single-cell, and spatial transcriptomics.
- DRAGEN Protein Quantification pipeline: Detect over 9.5k unique protein targets accurately with automated protein quantification, normalization, and quality control reporting, providing highly visual QC reports, making it easy to assess data quality and interpret results.
- DRAGEN 5-Base DNA pipeline: Detect methylated cytosines and DNA variants across the genome, unlocking multiomic insights from a single dataset with high fidelity. Uncover mechanisms that drive gene expression, cell fate, and disease progression with comprehensive methylation profiling.
- DRAGEN RNA pipeline: Quantify gene expression with high sensitivity and dynamic range using optimized alignment algorithms and hardware acceleration, enabling transcript-level resolution, differential expression analysis, and rapid interpretation of complex transcriptomic profiles.
- DRAGEN Single Cell RNA pipeline: Quantify transcripts and call variants in single-cell studies quickly through hardware acceleration that can process high-throughput data five times faster than the open source PIPseeker solution, making it ideal for large-scale studies.
- DRAGEN Spatial Transcriptome pipeline (in beta): Map gene expression in tissue context with precision using built-in error correction and barcode validation and automatically segment cells using custom-trained machine learning models.
Connected Multiomics also supports matrix files from other analysis solutions including Seurat, Scanpy, and 10X Cell Ranger and Space Ranger.
Biological interpretation
Connected Multiomics users can perform biological interpretation directly within the software using Correlation Engine integration. Correlation Engine provides access to over 26,000 multiomics studies and 100,000 reference databases (COSMIC, Gene Ontology, MSigDB, etc.), all curated and re-analyzed by DRAGEN for quality. This makes it one of the world’s largest biological databases.
- Automated correlation analysis: Proprietary algorithms scan billions of data points, finding comparable insights by matching private data to thousands of public datasets.
- Multiomic context: Correlation Engine enables cross-modality analysis—RNA, miRNA, protein, methylation, chromatin accessibility—so you can discover novel associations and validate biomarkers.
- Biologist-friendly tools: Guided workflows, push-button apps, and APIs make advanced analysis accessible to all users.
- Unprecedented discovery power: On average, querying a gene in Correlation Engine yields 145 times more significant omics data hits than PubMed, accelerating research and publication.
When combined with Connected Multiomics, Correlation Engine transforms raw multiomic data into actionable biological context, enabling discoveries that were previously impossible.
Additionally, Connected Multiomics offers access to KEGG (Kanehisa Laboratories) and Gene Set (UC San Diego and Broad Institute) databases.
Accelerating multiomic discovery with a unified, scalable platform
With Illumina Connected Multiomics, the future of biological discovery is here. By integrating proteomics, transcriptomics, epigenomics, and genomics data in one intuitive, cloud-based platform, researchers can finally break free from fragmented workflows and unlock deeper biological discoveries at scale. Whether you’re mapping cellular interactions, uncovering epigenetic mechanisms, or investigating potential links between diseases, Connected Multiomics delivers the tools to turn complex data into insights that drive scientific breakthroughs.
Ready to learn more or experience it for yourself?
Visit the product page | Watch demo videos | Request a free trial
__________________________
* Illumina DRAGEN pipelines have consistently ranked among the top in PrecisionFDA challenges for accuracy and speed. While these reports primarily focus on DNA and RNA, they validate DRAGEN’s core algorithms used across omics workflows. https://precision.fda.gov/challenges/13
** Early access product.
M-GL-03916 | For Research Use Only. Not for use in diagnostic procedures.