- Home
- News & Updates
- BaseSpace™ Sequence Hub now supports whole genome bisulfite sequencing (WGBS) data with Zymo-Seq Library Kit running on NovaSeq™ X series
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 06/06/2023
BaseSpace™ Sequence Hub now supports whole genome bisulfite sequencing (WGBS) data with Zymo-Seq Library Kit running on NovaSeq™ X series
Today, we’re thrilled to share NovaSeq X series data for whole genome bisulfite sequencing (WGBS) in partnership with Zymo Research. We are continuing a series of posts that shed more light on new run data publicly available on BaseSpace™ Sequence Hub (BSSH) for applications like whole genome bisulfite sequencing (WGBS).
WGBS is the gold standard for DNA methylation studies as it provides base-level methylation quantification for all cytosines. Zymo-Seq WGBS Library Kit was used in this experiment, in combination with Illumina DNA Prep library prep kit.
To prepare Zymo-Seq WGBS (D5465) libraries, intact genomic DNA is first bisulfite converted. Then, the following library preparation procedures are completed in a single tube: (1) second-strand synthesis, (2) tagmentation with Illumina DNA Prep library prep kit, and (3) library amplification and indexing (Figure 1).
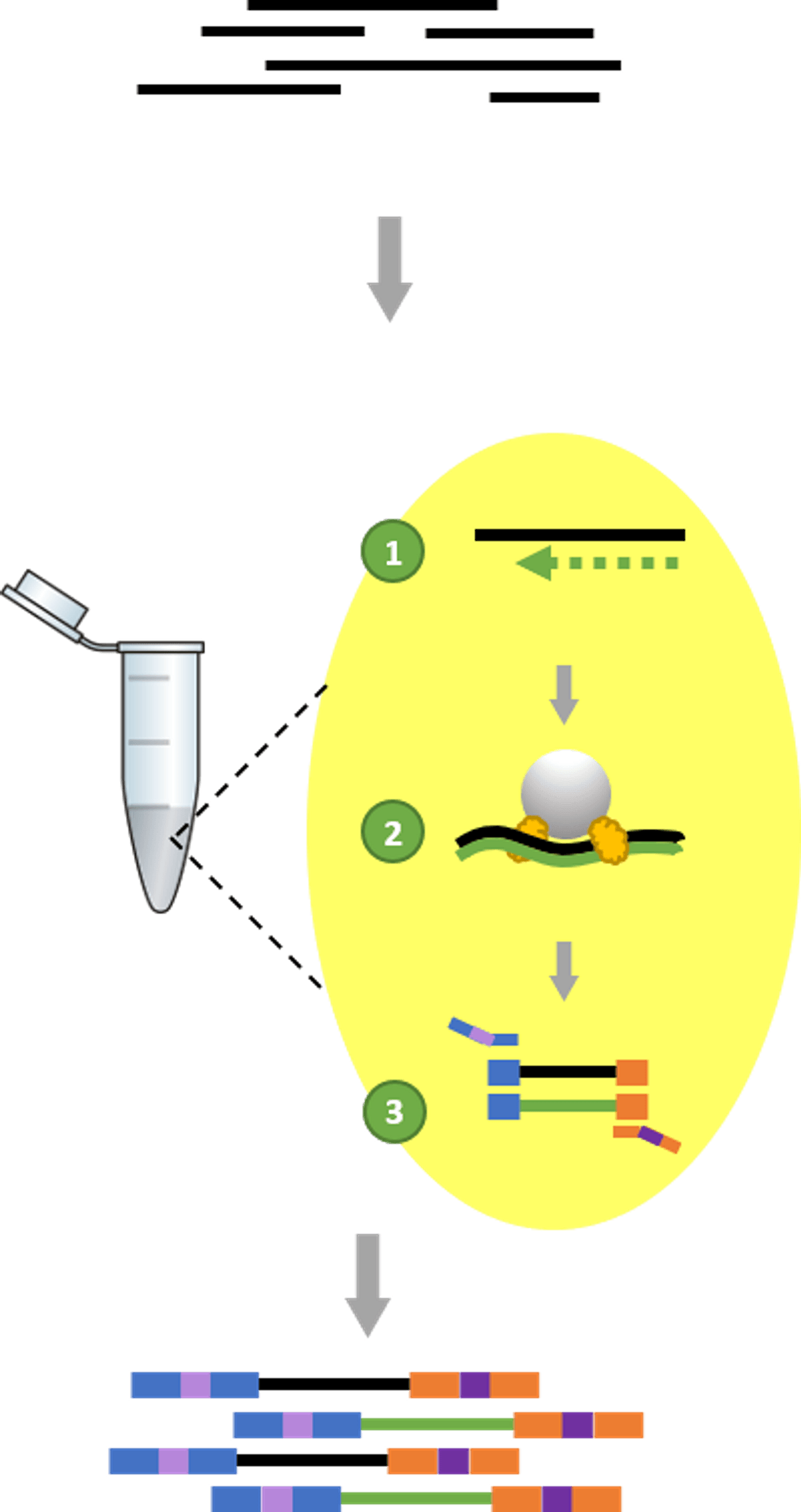
Figure 1: One Tube Library Prep Workflow using Zymo-Seq WGBS. Enzymatic reactions are consolidated in a single tube to minimize hands-on time.
After purification, libraries are ready for sequencing on Illumina instruments. Due to the necessary output, the libraries were sequenced on the NovaSeq X Plus. A 30x coverage per sample was targeted.
Sequencing on the NovaSeq X Plus
8 replicates each of matched human brain and spleen were prepared using the Zymo-Seq WGBS Library Kit A 0.5% unmethylated E.coli control was spiked in to assess the conversion.
Below are two links to directly import the runs and project folders into your BaseSpace account. These runs can be found under the “methyl-seq” category. Because these are public data sets, these are free and do not count against storage limits. See our blogpost to understand how to access these data.
You can use the demo data to compare with your own WGBS runs. See our first blog post in this series for additional details on how to evaluate your sequencing run quality.
Run link: https://ilmn-sso.basespace.illumina.com/s/0KFaBImFzUfD
Project link: https://ilmn-sso.basespace.illumina.com/s/ONRCldMlyFdB
Libraries were sequenced at 2x151 using NovaSeq X 10B flowcell. With Q30 = 92.60% and 3.47Tbp of yield, this WGBS run exceeds Illumina specifications (https://www.illumina.com/systems/sequencing-platforms/novaseq-x-plus/specifications.html).
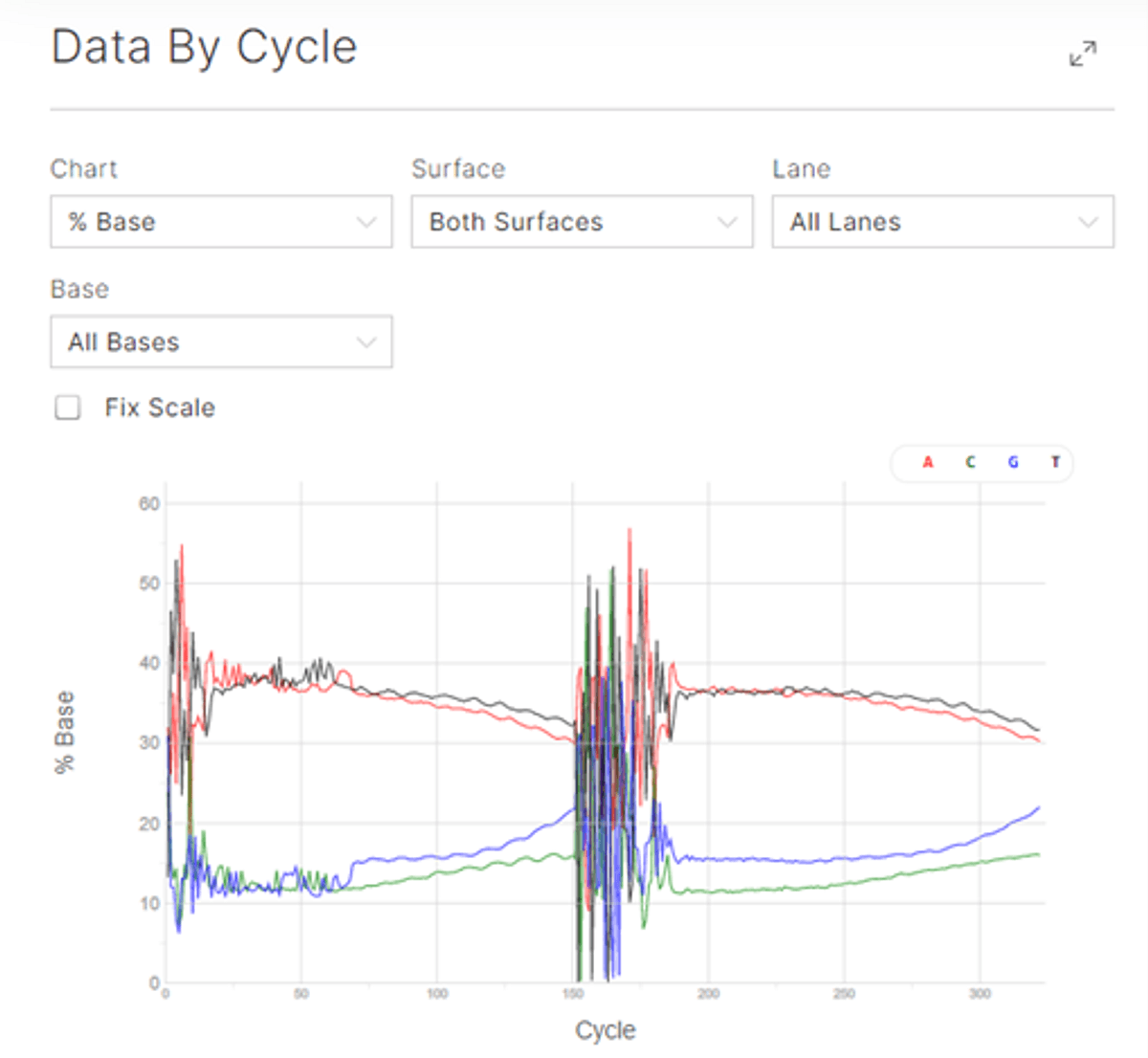
Figure 2: % base per cycle.
The data by cycle for the % base shows an unbalanced library – which is expected with the bisulfite conversion of cytosine. Illumina recommends to spike in at least 1% of PhiX – but it is better to spike in 5% or higher. Despite the unbalanced base composition, high Quality score is maintained on the NovaSeq X run.
Data analysis
Sequencing data were downsampled to 500M reads per sample for downstream analysis. Methylation analysis was performed using DRAGEN™ Methylation. Conversion efficiency was assessed with the unmethylated E. coli control, and was estimated to be higher than 99.5%.
Additional resources
NovaSeq 6000 data are also available for the WGBS Zymo-Seq data: https://developer.illumina.com/news-updates/whole-genome-bisulfite-sequencing-zymo-seq-data-now-available-on-illumina-basespace-sequence-hub
Support
Contact ZYMO Research for any questions regarding the bisulfite conversion, the library prep assay and the analysis results. Contact Illumina Tech support for sequencing and DRAGEN related questions.
Special thanks to our colleagues at ZYMO Research for providing ZYMO-seq reagents and
controls for this experiment, and hat tip to Illumina Scientist Kevin
Thai and the Systems Integration team for sequencing and analyzing these runs
for this collaboration.
For Research Use Only. Not for use in diagnostic procedures. M-GL-01847