- Home
- News & Updates
- Zymo Quick-16S Plus NGS Library Prep Kit (V3-V4) sequencing runs and data now available on Illumina BaseSpace™ Sequence Hub
-
BaseSpace™ Sequence Hub
-
Publications
-
News
- 12/02/2022
Zymo Quick-16S Plus NGS Library Prep Kit (V3-V4) sequencing runs and data now available on Illumina BaseSpace™ Sequence Hub
16S rRNA sequencing is a routine technique for microbiome composition profiling. Compared to shotgun metagenomics sequencing, 16S rRNA sequencing is more cost-effective and more robust; it generally requires less input DNA and is less impacted by the presence of host DNA.
Library preparation
16S libraries were generated with the Zymo Research Quick-16S Plus NGS Library Prep Kit (V3-V4) Quick-16S plus (D6420). The samples in this run are:
- ZymoBIOMICS Microbial Community DNA Standard (D6305)
- ZymoBIOMICS Fecal Reference with TruMatrix™ Technology (D6323)
- ZymoBIOMICS Gut Microbiome Standard (D6331)
- Fecal samples
Following the protocol guidelines, the final amplicon size after 1-Step PCR (targeted amplification and barcode addition) is ~606 bp.
NextSeq2000 sequencing
The 16S libraries were then sequenced on the NextSeq™ 2000 platform, using the newly launched NextSeq 1000/2000 P1 Reagent 600-cycle cartridge. Along with pooled 16S libraries, a 30% PhiX spike-in control was loaded on the cartridge for base calling error correction.
Run link: https://basespace.illumina.com...
These demo data are free to use on the BaseSpace Sequence Hub platform (see our previous post - https://developer.illumina.com/news-updates/demo-data-on-illumina-basespace-sequence-hub)
Evaluation of the sequencing run
To understand how to evaluate your sequencing run, see our previous blogpost "Does my sequencing run look good?"
With a Q30=91.5% and Yield around 84 Gb, this 16S run with Zymo libraries on NextSeq 2000 platform is meeting Illumina specifications in term of Q30 and yield.
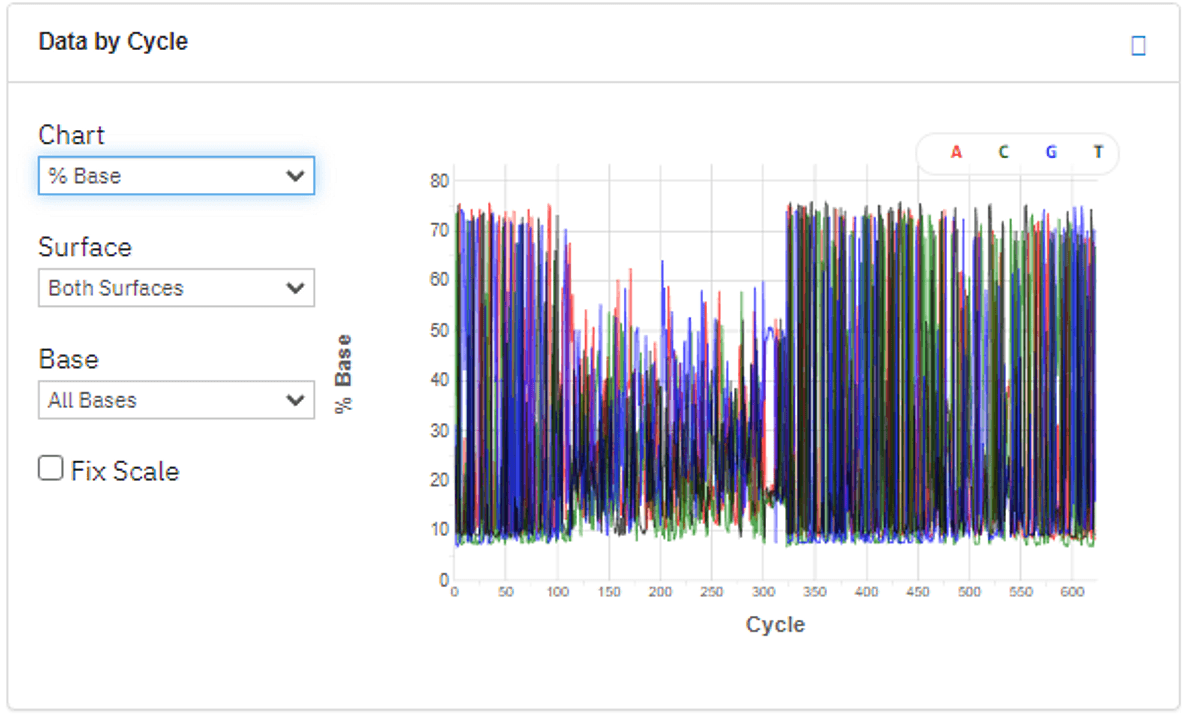
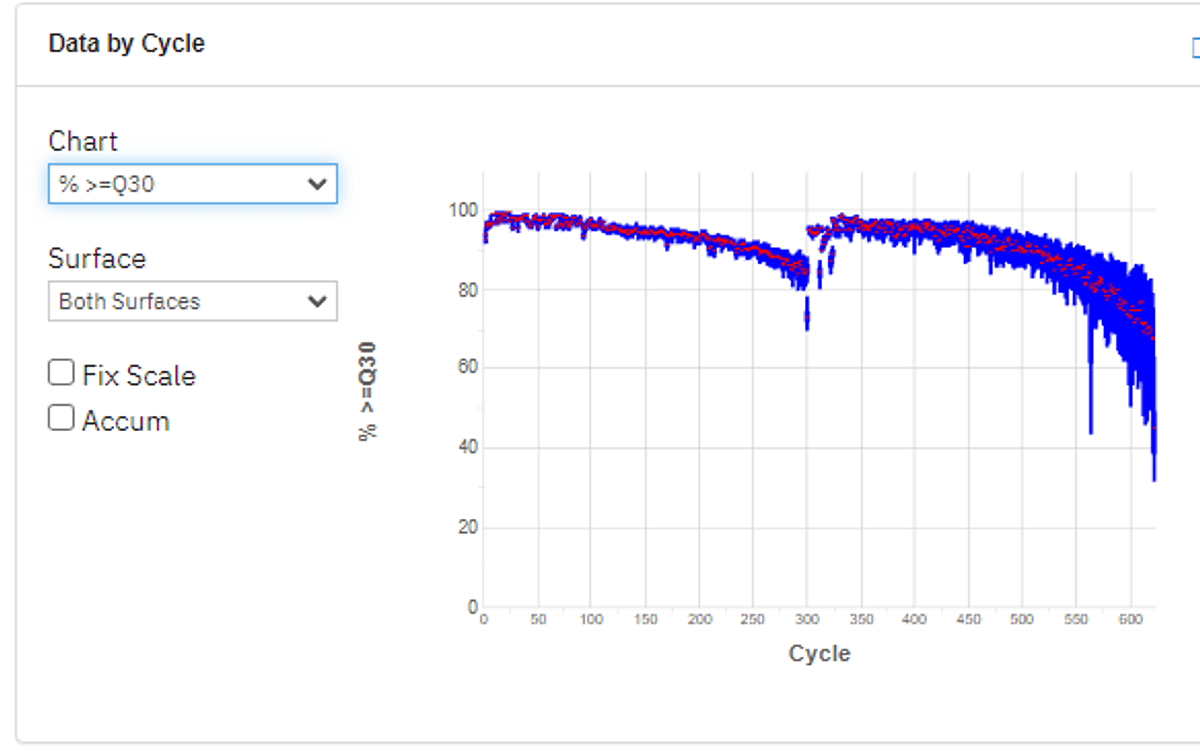
Figure 1: Percentage base composition, cycle by cycle (top) and Q30 chart (bottom). Despite the very low base diversity of the 16S libraries (left), high Q30 is maintained during the entire sequencing run.
Bioinformatics Analysis
ZymoBIOMICS® Targeted Sequencing Service was used for Microbiome Analysis. This report, in html format, is available under the BaseSpace “project” and can be downloaded.
Project link: https://basespace.illumina.com...
Unique amplicon sequences were inferred from raw reads using the Dada2 pipeline (Callahan et al., 2016). Chimeric sequences were also removed with the Dada2 pipeline. Taxonomy assignment was performed using Uclust from Qiime v.1.9.1. Taxonomy was assigned with the Zymo Research Database, a 16S database that is internally designed and curated, as reference.
Composition visualization, alpha-diversity, and beta-diversity analyses were performed with Qiime v.1.9.1 (Caporaso et al., 2010). If applicable, taxonomy that have significant abundance among different groups were identified by LEfSe (Segata et al., 2011) using default settings. Other analyses such as heatmaps, Taxa2SV_deomposer, and PCoA plots were performed with internal scripts.
Support
Contact ZYMO Research for any questions regarding the 16S library prep assay and the analysis results. Contact Illumina Tech support for sequencing related questions.
Special thanks to our colleagues at ZYMO Research for providing ZYMO-seq reagents and controls for this experiment and sharing their sequencing runs including downstream analysis for this collaboration.
For Research Use Only. Not for use in diagnostic procedures.