- Home
- News & Updates
- DRAGEN™ v4.0 - Powering Pharmacogenomics Insights with a New Star Allele Caller
-
DRAGEN
-
Product updates
- 09/20/2022
DRAGEN™ v4.0 - Powering Pharmacogenomics Insights with a New Star Allele Caller
The latest DRAGEN v4.0 release significantly expands pharmacogenomics (PGx) research capabilities, enabling accurate genotyping of complex genes.
Pharmacogenomics (PGx) is the study of how variations in the human genome dictate a person’s response to medications. In one study, more than 99% of people assessed had a genotype associated with a higher risk to at least one medication1. PGx allows healthcare providers to target more effective therapies, thereby reducing healthcare costs and better improving patient health. We have been continuously working to maximize the value of whole genome sequencing with DRAGEN, enabling you to get to a fully featured genome. With DRAGEN v4.0, you can further investigate complex PGx genes to determine the optimal genotype and corresponding metabolism status associated with the genotype for 20 Tier 12 PGx genes. DRAGEN v4.0 PGx research capabilities provide powerful insights such as identification of individual genotypes and gene expression associated with optimal metabolism.
Let’s dive into new DRAGEN PGx capabilities:
New PGx Star Allele Calling
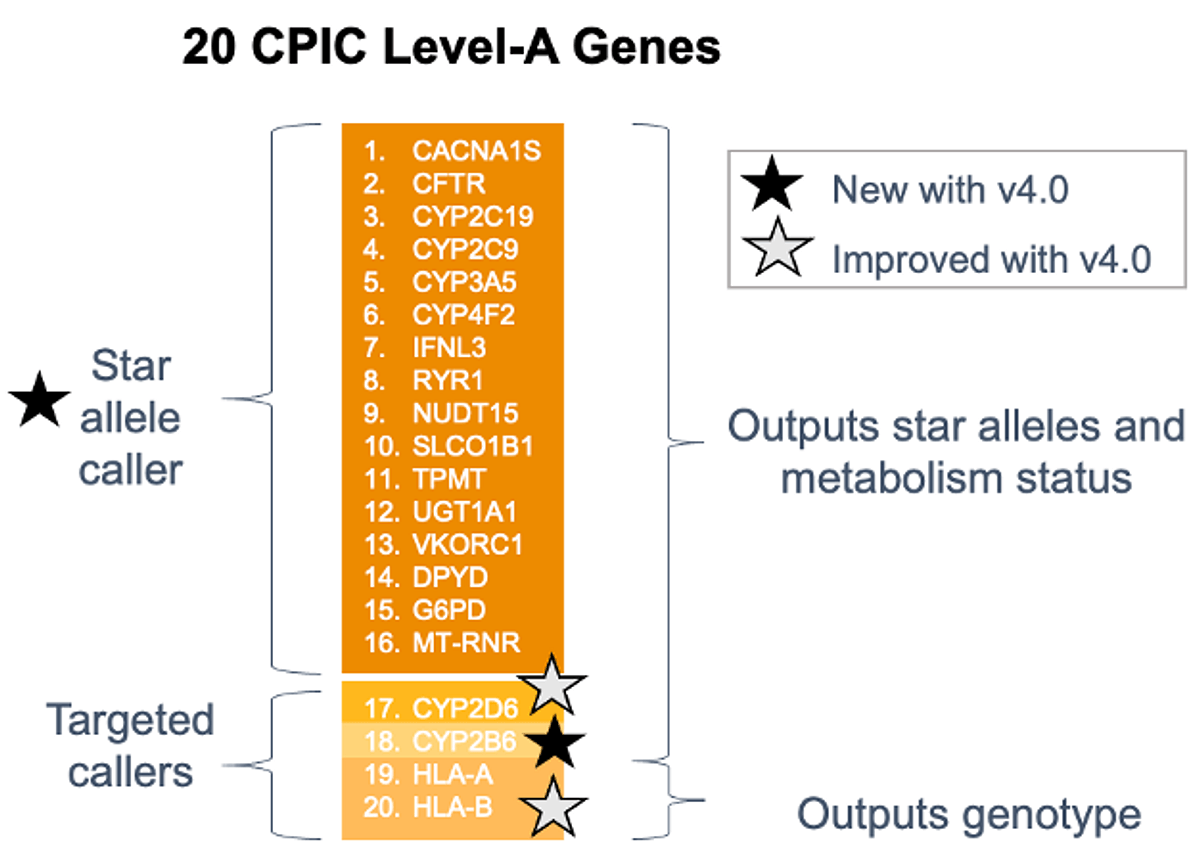
One of the PGx research capabilities new to DRAGEN v4.0 is the Star Allele Caller, which calls star alleles and metabolism status for 16 complex, Tier 1 genes. These genes have been assigned Level-A guidelines for gene-drug interactions by CPIC. Targeted callers, which we’ll go over in more depth later, call four other Tier 1 genes. Star alleles are haplotypes of a gene characterized by specific variants that dictate the drug metabolism status of the gene. DRAGEN star allele caller identifies the genotypes of the following 16 Tier-1 PGx genes: CACNA1S, CFTR, CYP2C19, CYP2C9, CYP3A5, CYP4F2, IFNL3, RYR1, NUDT15, SLCO1B1, TPMT, UGT1A1, VKORC1, DPYD, G6PD, MT-RNR1. For example, someone with a deficiency in the G6PD gene may have increased risks of adverse reactions to drugs such as primaquine (antibiotic), chloroquine (antiparasitic), dapsone (antiparasitic), and rasburicase (a drug used before/after chemotherapy). For each gene, the caller returns the optimal genotype and the corresponding metabolism status associated with that genotype.
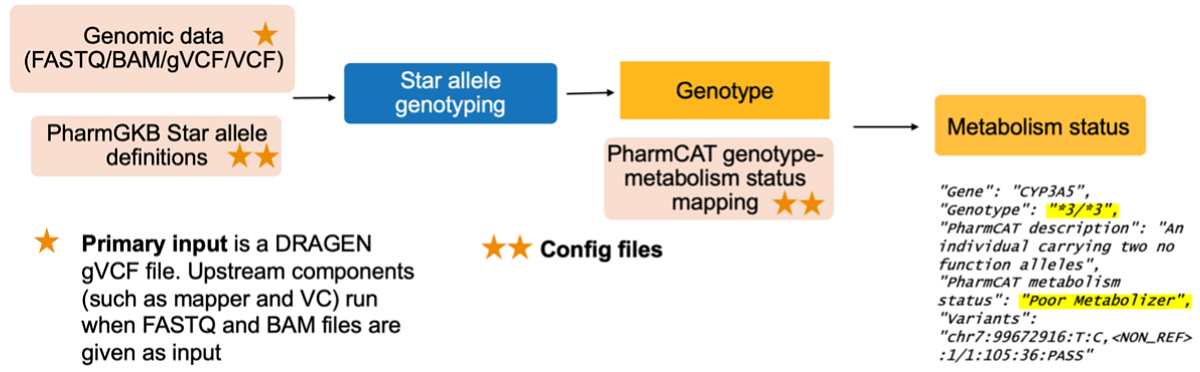
The Star Allele Caller works by starting with genomic data in the form of a FASTQ/BAM/gVCF/VCF file. The primary input is a DRAGEN gVCF file. Upstream components, such as mapper and VC, run when FASTQ and BAM files are given as input. You can also start the workflow with PharmGKB Star allele definitions, which are configuration files. From there, star allele genotyping commences and PharmCAT genotype-metabolism status mapping (config file) produces a genotype. The result of the Star Allele Caller pipeline is an output of metabolism status.
The Star Allele Caller workflow:
Enhanced PGx Targeted Calling
In DRAGEN v3.7, we introduced targeted calling for CYP2D6. DRAGEN v4.0 now also enables targeting calling for CYP2B6, an important but notoriously difficult-to-call pharmacogene of the cytochrome P450 family.
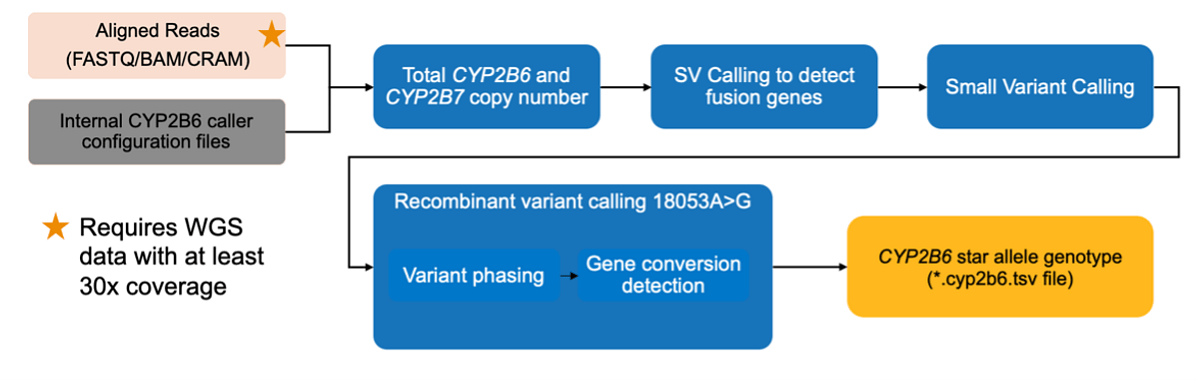
Gene CYP2B6 plays a role in the metabolism of efavirenz, bupropion, and cyclophosphamide and is also expressed in the brain, where it may be an important factor in the metabolism of CNS-acting drugs and neurological side-effects of drug treatments. Detecting variants in CYP2B6 is complicated by sequence homology with its pseudogene paralog CYP2B7. By differentiating between reads mapping to the CYP2B6 gene and its paralog, the new DRAGEN CYP2B6 caller can detect 37 different star alleles known in the population. The CYP2B6 Caller works by starting with aligned reads in the form of a FASTQ/BAM/CRAM file. Please note, this requires WGS data with at least 30x coverage. You can also start the CYP2B6 Caller workflow with internal CYP2B6 caller configuration files. From there, total CYP2B6 and CYP2B7 copy number are extracted. Then, small variant calling is used to detect fusion genes. The next step is called recombinant variant calling (18053A>G) and includes variant phasing and subsequent gene conversion detection. The result of this process is a CYP2B6 star allele genotype in the form of a *.cyp2b6.tsv file. DRAGEN CYP2B6 caller can be optionally enabled as part of the Germline pipeline and supports reference genomes GRCh37, hg19, and hg28.
The CYP2B6 Targeted Caller workflow:

DRAGEN v4.0 brings a new HLA genotyping algorithm as well, expanding research capabilities to include a nucleotide-based HLA-specific alignment and improved probabilistic modeling inferring HLA genotypes. This includes improved HLA typing accuracy (WGS, WES > 99% accuracy; TSO500 panels > 97% accuracy). HLA, which stands for human leukocyte antigen, is a highly polymorphic region consisting of multiple genes involved in immune regulation. HLA typing is used to target recurrent mutations implicated in cancer pathogenesis and to match patients and donors for bone marrow or cord blood transplants. Genotyping HLA genes is notoriously difficult due to high levels of sequence homology between HLA genes and pseudogenes and dense variability. Formerly, genotyping relied on amino-acid alignment with maximal exact-match and integer linear programming. DRAGEN’s new HLA component performs nucleotide-based alignment and expectation-maximization for genotyping, leveraging DRAGEN map-align with HLA-specific reference from IMGT/HLA database (v3.45). The new DRAGEN HLA genotyping algorithm corrects miscalls due to incorrect translation and requires no additional command-line options. The new and improved HLA workflow completes nucleotide-based alignments using DRAGEN map-align and expectation-maximization for genotyping.
HLA workflow:
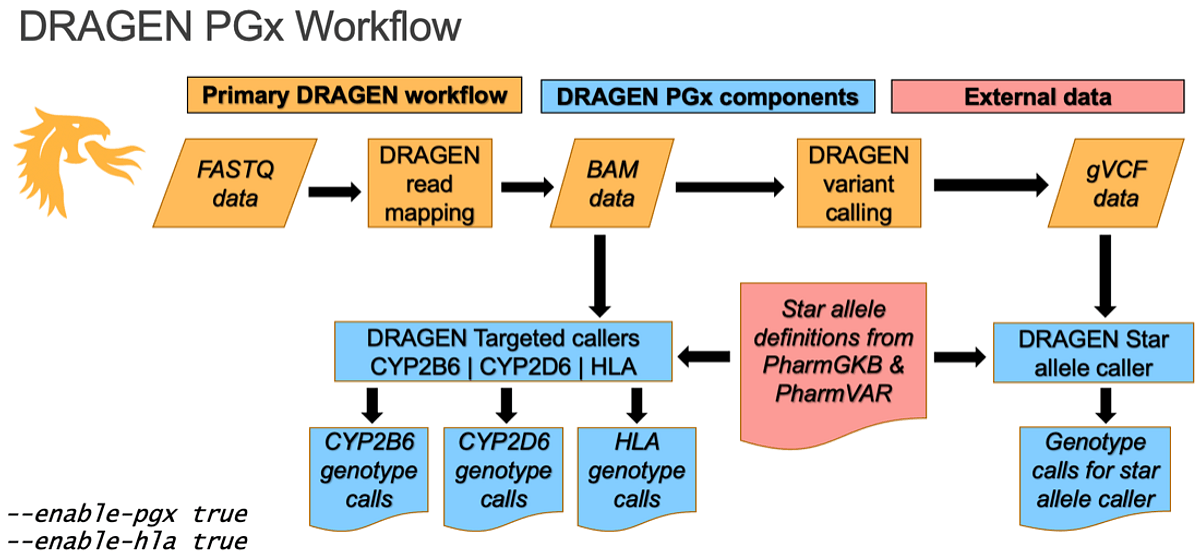
The new DRAGEN v4.0 PGx workflow at a glance:
Summary:
DRAGEN v4.0 has brought several exciting new PGx research capabilities to our customers. These include:
- Star allele caller, which calls star alleles and metabolism status for 16 PGx genes
- Targeted callers, which call star alleles and metabolism status for difficult genes CYP2D6 and CYP2B6
- Targeted HLA caller, which outputs the genotype
Learn More:
For more examples of how the DRAGEN 4.0 release is full of advances in comprehensiveness, accuracy, and efficiency, read our DRAGEN v4.0 release blog
For an in-depth summary of all DRAGEN 4.0 features, view our user guide.
Visit the DRAGEN page to learn more about our technology and other updates, including our latest v4 server release.
Other Resources:
- Reisberg S et al. Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions. Genet Med. 2019 Jun;21(6):1345-1354.
- *Tier 1 = CPIC Level-A genes as of March 1, 2022
For Research Use Only. Not for use in diagnostic procedures.